Supercomputer Significantly Shown to Speed up Genome Analysis
|
By LabMedica International staff writers Posted on 06 Mar 2014 |

Image: The Beagle computer at Argonne National Laboratory, one of the world’s fastest supercomputers designed for life sciences, is able to analyze 240 full genomes in about two days (Photo courtesy of the University of Chicago Medicine).
Investigators, working with one of the world’s fastest supercomputers designed for life sciences, recently reported that genome analysis could now be drastically accelerated.
The supercomputer named Beagle is based at Argonne US National Laboratory (Argonne, IL, USA), and it is able to analyze 240 full genomes in about two days. Although the time and cost of sequencing a complete human genome has nosedived, analyzing the resulting three billion base pairs of genetic data from only one genome can take many months.
University of Chicago (IL, USA) scientists working with the Beagle supercomputer published their findings online on February 12, 2014, in the journal Bioinformatics. “This is a resource that can change patient management and, over time, add depth to our understanding of the genetic causes of risk and disease,” said study author Elizabeth McNally, MD, PhD, a professor of medicine and human genetics and director of the Cardiovascular Genetics clinic at the University of Chicago Medicine.
“The supercomputer can process many genomes simultaneously rather than one at a time,” said first author Megan Puckelwartz, a graduate student in McNally’s laboratory. “It converts whole genome sequencing, which has primarily been used as a research tool, into something that is immediately valuable for patient care.”
Because the genome is so huge, those investigators involved in clinical genetics have turned to exome sequencing, which focuses on the 2% or less of the genome that codes for proteins. This application is frequently helpful because an estimated 85% of disease-causing mutations are located in coding regions. However, the rest, approximately 15% of clinically significant mutations, come from noncoding regions, once referred to as “junk DNA,” but now known to serve important functions. If not for the vast data-processing analysis challenges, whole genome sequencing would be the method of choice.
To evaluate the system, the scientists utilized raw sequencing data from 61 human genomes and analyzed that data on Beagle. They used publicly available software packages and 25% of the computer’s total capacity. They discovered that shifting to the supercomputer setting improved accuracy and dramatically accelerated speed. “Improving analysis through both speed and accuracy reduces the price per genome,” Dr. McNally said. “With this approach, the price for analyzing an entire genome is less than the cost of the looking at just a fraction of genome. New technology promises to bring the costs of sequencing down to around USD 1,000 per genome. Our goal is get the cost of analysis down into that range.”
“This work vividly demonstrates the benefits of dedicating a powerful supercomputer resource to biomedical research,” said co-author Dr. Ian Foster, director of the Computation Institute and a professor of computer science. “The methods developed here will be instrumental in relieving the data analysis bottleneck that researchers face as genetic sequencing grows cheaper and faster.”
The finding has immediate medical applications. Dr. McNally’s Cardiovascular Genetics clinic, for example, relies on comprehensive cross-examination of the genes from an initial patient as well as a number of family members to understand, treat, and prevent disease. More than 50 genes can contribute to cardiomyopathy. Other genes can stimulate rhythm disorders, heart failure, or vascular problems. “We start genetic testing with the patient,” she said, “but when we find a significant mutation we have to think about testing the whole family to identify individuals at risk.”
The range of testable mutations has greatly expanded. “In the early days we would test one to three genes,” Dr. McNally said. “In 2007, we did our first five-gene panel. Now we order 50 to 70 genes at a time, which usually gets us an answer. At that point, it can be more useful and less expensive to sequence the whole genome.”
These genomic data combined with meticulous attention to patient and family histories “adds to our knowledge about these inherited disorders,” Dr. McNally said. “It can refine the classification of these disorders. By paying close attention to family members with genes that place then at increased risk, but who do not yet show signs of disease, we can investigate early phases of a disorder. In this setting, each patient is a big-data problem.”
Beagle, a Cray XE6 supercomputer housed in the Theory and Computing Sciences (TCS) building at Argonne National Laboratory, supports computation, simulation, and data analysis for the biomedical research community. It was named after the HMS Beagle, the ship that carried Charles Darwin on his celebrated scientific voyage in 1831.
Related Links:
Argonne U.S. National Laboratory
University of Chicago
The supercomputer named Beagle is based at Argonne US National Laboratory (Argonne, IL, USA), and it is able to analyze 240 full genomes in about two days. Although the time and cost of sequencing a complete human genome has nosedived, analyzing the resulting three billion base pairs of genetic data from only one genome can take many months.
University of Chicago (IL, USA) scientists working with the Beagle supercomputer published their findings online on February 12, 2014, in the journal Bioinformatics. “This is a resource that can change patient management and, over time, add depth to our understanding of the genetic causes of risk and disease,” said study author Elizabeth McNally, MD, PhD, a professor of medicine and human genetics and director of the Cardiovascular Genetics clinic at the University of Chicago Medicine.
“The supercomputer can process many genomes simultaneously rather than one at a time,” said first author Megan Puckelwartz, a graduate student in McNally’s laboratory. “It converts whole genome sequencing, which has primarily been used as a research tool, into something that is immediately valuable for patient care.”
Because the genome is so huge, those investigators involved in clinical genetics have turned to exome sequencing, which focuses on the 2% or less of the genome that codes for proteins. This application is frequently helpful because an estimated 85% of disease-causing mutations are located in coding regions. However, the rest, approximately 15% of clinically significant mutations, come from noncoding regions, once referred to as “junk DNA,” but now known to serve important functions. If not for the vast data-processing analysis challenges, whole genome sequencing would be the method of choice.
To evaluate the system, the scientists utilized raw sequencing data from 61 human genomes and analyzed that data on Beagle. They used publicly available software packages and 25% of the computer’s total capacity. They discovered that shifting to the supercomputer setting improved accuracy and dramatically accelerated speed. “Improving analysis through both speed and accuracy reduces the price per genome,” Dr. McNally said. “With this approach, the price for analyzing an entire genome is less than the cost of the looking at just a fraction of genome. New technology promises to bring the costs of sequencing down to around USD 1,000 per genome. Our goal is get the cost of analysis down into that range.”
“This work vividly demonstrates the benefits of dedicating a powerful supercomputer resource to biomedical research,” said co-author Dr. Ian Foster, director of the Computation Institute and a professor of computer science. “The methods developed here will be instrumental in relieving the data analysis bottleneck that researchers face as genetic sequencing grows cheaper and faster.”
The finding has immediate medical applications. Dr. McNally’s Cardiovascular Genetics clinic, for example, relies on comprehensive cross-examination of the genes from an initial patient as well as a number of family members to understand, treat, and prevent disease. More than 50 genes can contribute to cardiomyopathy. Other genes can stimulate rhythm disorders, heart failure, or vascular problems. “We start genetic testing with the patient,” she said, “but when we find a significant mutation we have to think about testing the whole family to identify individuals at risk.”
The range of testable mutations has greatly expanded. “In the early days we would test one to three genes,” Dr. McNally said. “In 2007, we did our first five-gene panel. Now we order 50 to 70 genes at a time, which usually gets us an answer. At that point, it can be more useful and less expensive to sequence the whole genome.”
These genomic data combined with meticulous attention to patient and family histories “adds to our knowledge about these inherited disorders,” Dr. McNally said. “It can refine the classification of these disorders. By paying close attention to family members with genes that place then at increased risk, but who do not yet show signs of disease, we can investigate early phases of a disorder. In this setting, each patient is a big-data problem.”
Beagle, a Cray XE6 supercomputer housed in the Theory and Computing Sciences (TCS) building at Argonne National Laboratory, supports computation, simulation, and data analysis for the biomedical research community. It was named after the HMS Beagle, the ship that carried Charles Darwin on his celebrated scientific voyage in 1831.
Related Links:
Argonne U.S. National Laboratory
University of Chicago
Latest BioResearch News
- Genome Analysis Predicts Likelihood of Neurodisability in Oxygen-Deprived Newborns
- Gene Panel Predicts Disease Progession for Patients with B-cell Lymphoma
- New Method Simplifies Preparation of Tumor Genomic DNA Libraries
- New Tool Developed for Diagnosis of Chronic HBV Infection
- Panel of Genetic Loci Accurately Predicts Risk of Developing Gout
- Disrupted TGFB Signaling Linked to Increased Cancer-Related Bacteria
- Gene Fusion Protein Proposed as Prostate Cancer Biomarker
- NIV Test to Diagnose and Monitor Vascular Complications in Diabetes
- Semen Exosome MicroRNA Proves Biomarker for Prostate Cancer
- Genetic Loci Link Plasma Lipid Levels to CVD Risk
- Newly Identified Gene Network Aids in Early Diagnosis of Autism Spectrum Disorder
- Link Confirmed between Living in Poverty and Developing Diseases
- Genomic Study Identifies Kidney Disease Loci in Type I Diabetes Patients
- Liquid Biopsy More Effective for Analyzing Tumor Drug Resistance Mutations
- New Liquid Biopsy Assay Reveals Host-Pathogen Interactions
- Method Developed for Enriching Trophoblast Population in Samples
Channels
Clinical Chemistry
view channel
New PSA-Based Prognostic Model Improves Prostate Cancer Risk Assessment
Prostate cancer is the second-leading cause of cancer death among American men, and about one in eight will be diagnosed in their lifetime. Screening relies on blood levels of prostate-specific antigen... Read more
Extracellular Vesicles Linked to Heart Failure Risk in CKD Patients
Chronic kidney disease (CKD) affects more than 1 in 7 Americans and is strongly associated with cardiovascular complications, which account for more than half of deaths among people with CKD.... Read moreMolecular Diagnostics
view channel
Diagnostic Device Predicts Treatment Response for Brain Tumors Via Blood Test
Glioblastoma is one of the deadliest forms of brain cancer, largely because doctors have no reliable way to determine whether treatments are working in real time. Assessing therapeutic response currently... Read more
Blood Test Detects Early-Stage Cancers by Measuring Epigenetic Instability
Early-stage cancers are notoriously difficult to detect because molecular changes are subtle and often missed by existing screening tools. Many liquid biopsies rely on measuring absolute DNA methylation... Read more
“Lab-On-A-Disc” Device Paves Way for More Automated Liquid Biopsies
Extracellular vesicles (EVs) are tiny particles released by cells into the bloodstream that carry molecular information about a cell’s condition, including whether it is cancerous. However, EVs are highly... Read more
Blood Test Identifies Inflammatory Breast Cancer Patients at Increased Risk of Brain Metastasis
Brain metastasis is a frequent and devastating complication in patients with inflammatory breast cancer, an aggressive subtype with limited treatment options. Despite its high incidence, the biological... Read moreHematology
view channel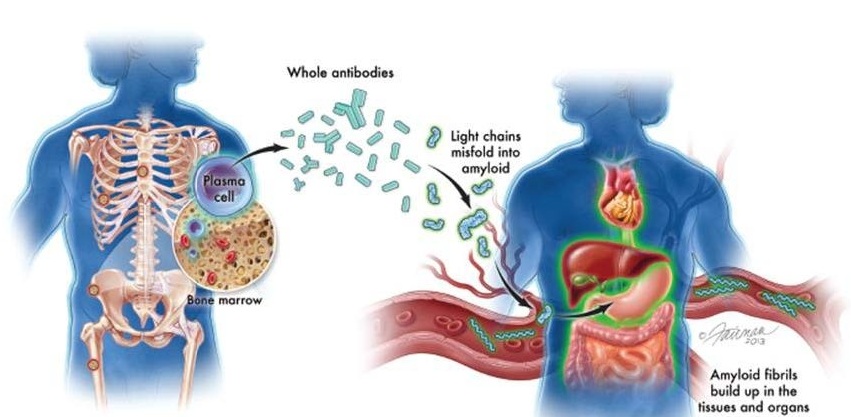
New Guidelines Aim to Improve AL Amyloidosis Diagnosis
Light chain (AL) amyloidosis is a rare, life-threatening bone marrow disorder in which abnormal amyloid proteins accumulate in organs. Approximately 3,260 people in the United States are diagnosed... Read more
Fast and Easy Test Could Revolutionize Blood Transfusions
Blood transfusions are a cornerstone of modern medicine, yet red blood cells can deteriorate quietly while sitting in cold storage for weeks. Although blood units have a fixed expiration date, cells from... Read more
Automated Hemostasis System Helps Labs of All Sizes Optimize Workflow
High-volume hemostasis sections must sustain rapid turnaround while managing reruns and reflex testing. Manual tube handling and preanalytical checks can strain staff time and increase opportunities for error.... Read more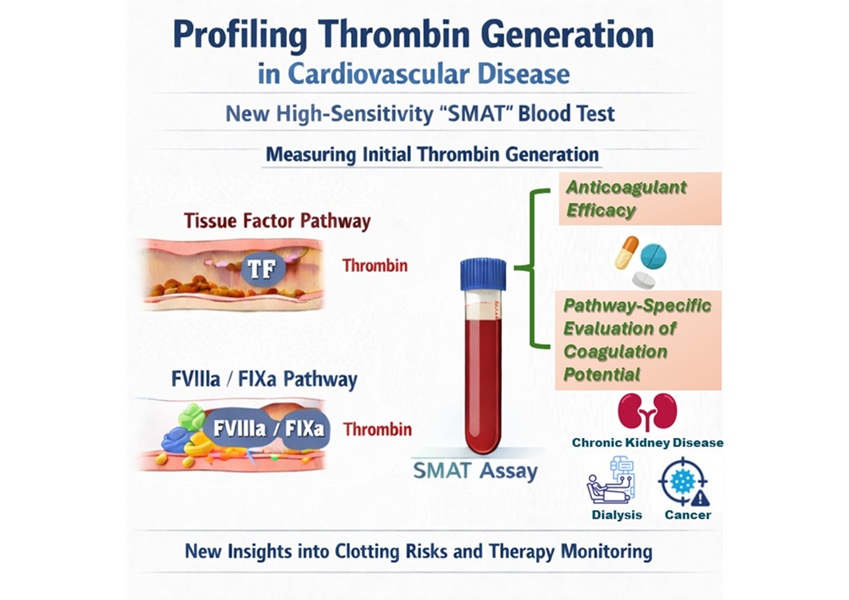
High-Sensitivity Blood Test Improves Assessment of Clotting Risk in Heart Disease Patients
Blood clotting is essential for preventing bleeding, but even small imbalances can lead to serious conditions such as thrombosis or dangerous hemorrhage. In cardiovascular disease, clinicians often struggle... Read moreImmunology
view channelBlood Test Identifies Lung Cancer Patients Who Can Benefit from Immunotherapy Drug
Small cell lung cancer (SCLC) is an aggressive disease with limited treatment options, and even newly approved immunotherapies do not benefit all patients. While immunotherapy can extend survival for some,... Read more
Whole-Genome Sequencing Approach Identifies Cancer Patients Benefitting From PARP-Inhibitor Treatment
Targeted cancer therapies such as PARP inhibitors can be highly effective, but only for patients whose tumors carry specific DNA repair defects. Identifying these patients accurately remains challenging,... Read more
Ultrasensitive Liquid Biopsy Demonstrates Efficacy in Predicting Immunotherapy Response
Immunotherapy has transformed cancer treatment, but only a small proportion of patients experience lasting benefit, with response rates often remaining between 10% and 20%. Clinicians currently lack reliable... Read moreMicrobiology
view channel
Comprehensive Review Identifies Gut Microbiome Signatures Associated With Alzheimer’s Disease
Alzheimer’s disease affects approximately 6.7 million people in the United States and nearly 50 million worldwide, yet early cognitive decline remains difficult to characterize. Increasing evidence suggests... Read moreAI-Powered Platform Enables Rapid Detection of Drug-Resistant C. Auris Pathogens
Infections caused by the pathogenic yeast Candida auris pose a significant threat to hospitalized patients, particularly those with weakened immune systems or those who have invasive medical devices.... Read morePathology
view channel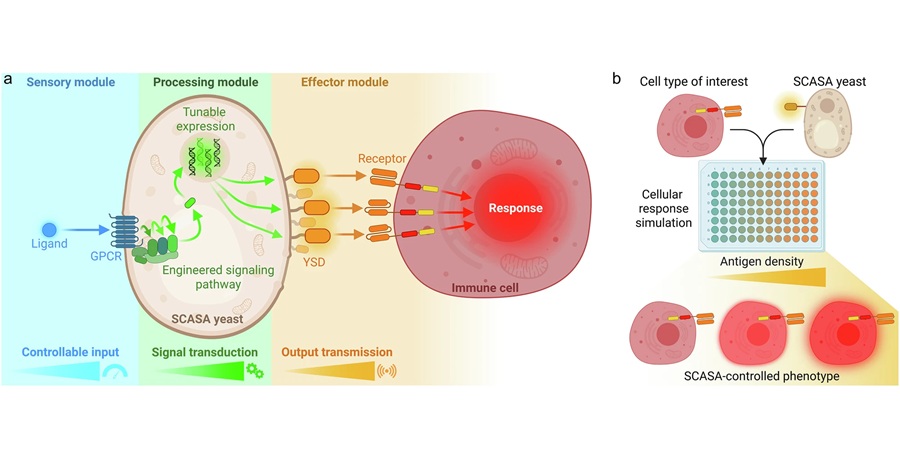
Engineered Yeast Cells Enable Rapid Testing of Cancer Immunotherapy
Developing new cancer immunotherapies is a slow, costly, and high-risk process, particularly for CAR T cell treatments that must precisely recognize cancer-specific antigens. Small differences in tumor... Read more
First-Of-Its-Kind Test Identifies Autism Risk at Birth
Autism spectrum disorder is treatable, and extensive research shows that early intervention can significantly improve cognitive, social, and behavioral outcomes. Yet in the United States, the average age... Read moreTechnology
view channel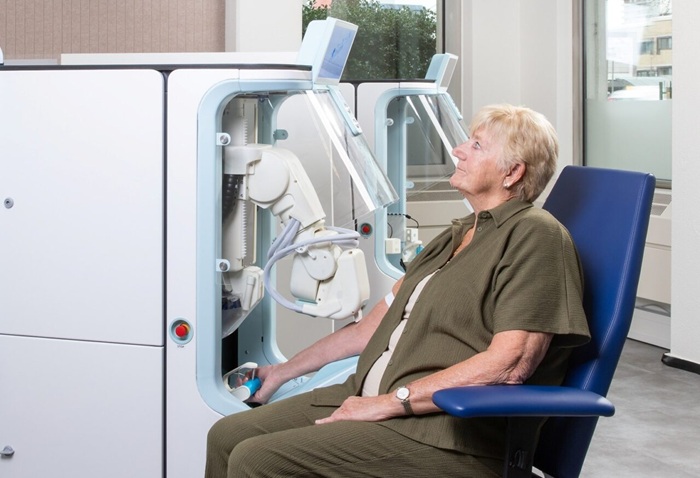
Robotic Technology Unveiled for Automated Diagnostic Blood Draws
Routine diagnostic blood collection is a high‑volume task that can strain staffing and introduce human‑dependent variability, with downstream implications for sample quality and patient experience.... Read more
ADLM Launches First-of-Its-Kind Data Science Program for Laboratory Medicine Professionals
Clinical laboratories generate billions of test results each year, creating a treasure trove of data with the potential to support more personalized testing, improve operational efficiency, and enhance patient care.... Read moreAptamer Biosensor Technology to Transform Virus Detection
Rapid and reliable virus detection is essential for controlling outbreaks, from seasonal influenza to global pandemics such as COVID-19. Conventional diagnostic methods, including cell culture, antigen... Read more
AI Models Could Predict Pre-Eclampsia and Anemia Earlier Using Routine Blood Tests
Pre-eclampsia and anemia are major contributors to maternal and child mortality worldwide, together accounting for more than half a million deaths each year and leaving millions with long-term health complications.... Read moreIndustry
view channelNew Collaboration Brings Automated Mass Spectrometry to Routine Laboratory Testing
Mass spectrometry is a powerful analytical technique that identifies and quantifies molecules based on their mass and electrical charge. Its high selectivity, sensitivity, and accuracy make it indispensable... Read more
AI-Powered Cervical Cancer Test Set for Major Rollout in Latin America
Noul Co., a Korean company specializing in AI-based blood and cancer diagnostics, announced it will supply its intelligence (AI)-based miLab CER cervical cancer diagnostic solution to Mexico under a multi‑year... Read more
Diasorin and Fisher Scientific Enter into US Distribution Agreement for Molecular POC Platform
Diasorin (Saluggia, Italy) has entered into an exclusive distribution agreement with Fisher Scientific, part of Thermo Fisher Scientific (Waltham, MA, USA), for the LIAISON NES molecular point-of-care... Read more