Liquid Biopsy More Effective for Analyzing Tumor Drug Resistance Mutations
By LabMedica International staff writers
Posted on 16 Sep 2019
Results published in a recent paper suggested that liquid biopsy – specifically analysis of cell-free tumor DNA (ctDNA) – may capture the genetic diversity of acquired drug resistance better than classical surgical biopsy of the tumor.Posted on 16 Sep 2019
The effectiveness of ctDNA versus standard single-lesion tumor biopsies has not been directly compared in larger-scale prospective cohorts of patients receiving targeted therapy. Therefore, investigators at the Broad Institute of MIT and Harvard (Boston MA, USA) and their colleagues undertook a prospective study of a cohort of 42 patients with molecularly defined gastrointestinal cancers and acquired resistance to targeted therapy.This study was included in a five-year collaboration that analyzed tumors before and after the onset of drug resistance.
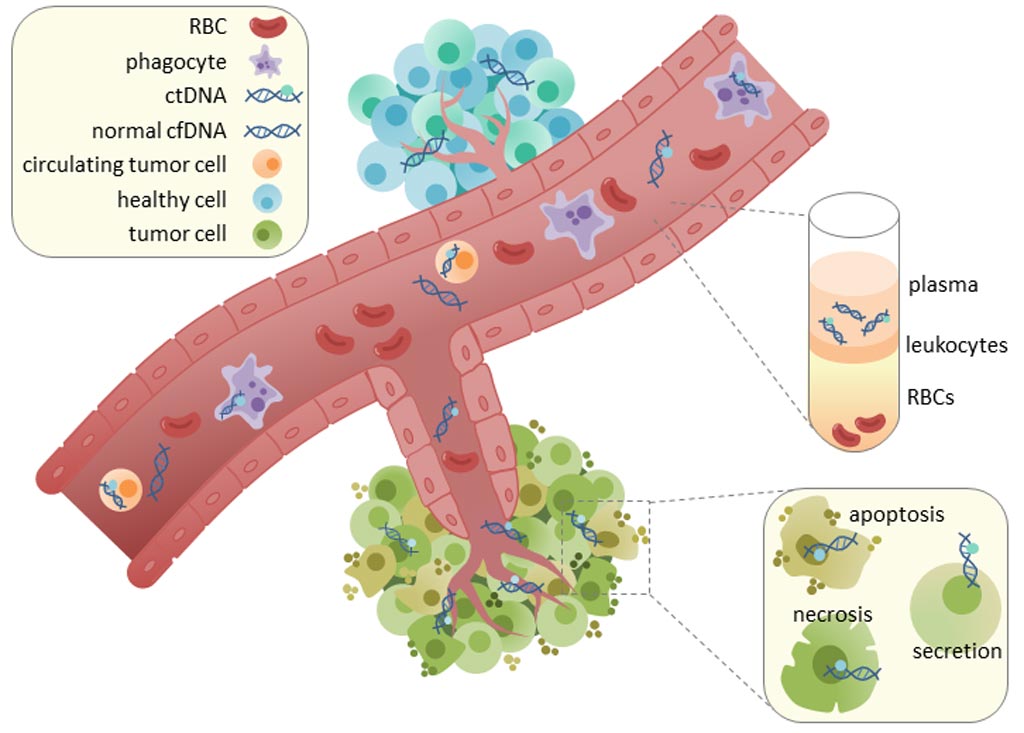
Image: Circulating tumor cell-free DNA (ctDNA) is found in serum and plasma fractions from blood. The mechanism of ctDNA release is unknown, though apoptosis, necrosis, and active secretion from tumor cells have been hypothesized (Photo courtesy of Wikimedia Commons).
Tumors were analyzed using both liquid and tissue biopsies procedures. The investigators employed a suite of computational tools developed at the Broad Institute, known as PhylogicNDT, to analyze tumor DNA and resistance mutations.
Direct comparison of ctDNA versus tumor biopsy revealed that ctDNA more frequently identified clinically relevant resistance alterations and multiple resistance mechanisms, detecting resistance alterations not found in the matched tumor biopsy in 78% of cases. Whole-exome sequencing of serial ctDNA, tumor biopsies and rapid autopsy specimens elucidated substantial geographic and evolutionary differences across separate tumors.
"This study is the largest to date to directly compare liquid biopsy to tumor biopsy in the setting of cancer resistance," said senior author Dr. Ryan Corcoran, assistant professor of medicine at Harvard Medical School. "Our findings suggest that liquid biopsy may be the preferred clinical modality for assessing how patients' tumors have evolved after they have become resistant to therapy."
The investigators proposed that the data collected in this study indicated that acquired resistance was frequently characterized by profound tumor heterogeneity, and that the emergence of multiple resistance alterations in an individual patient may represent the “rule” rather than the “exception.”
The study was published in the September 9, 2019, online edition of the journal Nature Medicine.
Related Links:
Broad Institute