New Liquid Biopsy Assay Reveals Host-Pathogen Interactions
By LabMedica International staff writers
Posted on 09 Sep 2019
A sensitive new assay employs high-throughput DNA sequencing to identify a large range of viral and bacterial pathogens in clinical samples, distinguish them from host DNA, and determine the degree of host tissue injury due to interaction with the pathogens.Posted on 09 Sep 2019
While high-throughput DNA sequencing offers an unbiased approach to identify pathogens in clinical samples, this method does not take into account information about the host, which is often critical to distinguish infection from infectious disease, and to assess the severity of disease.
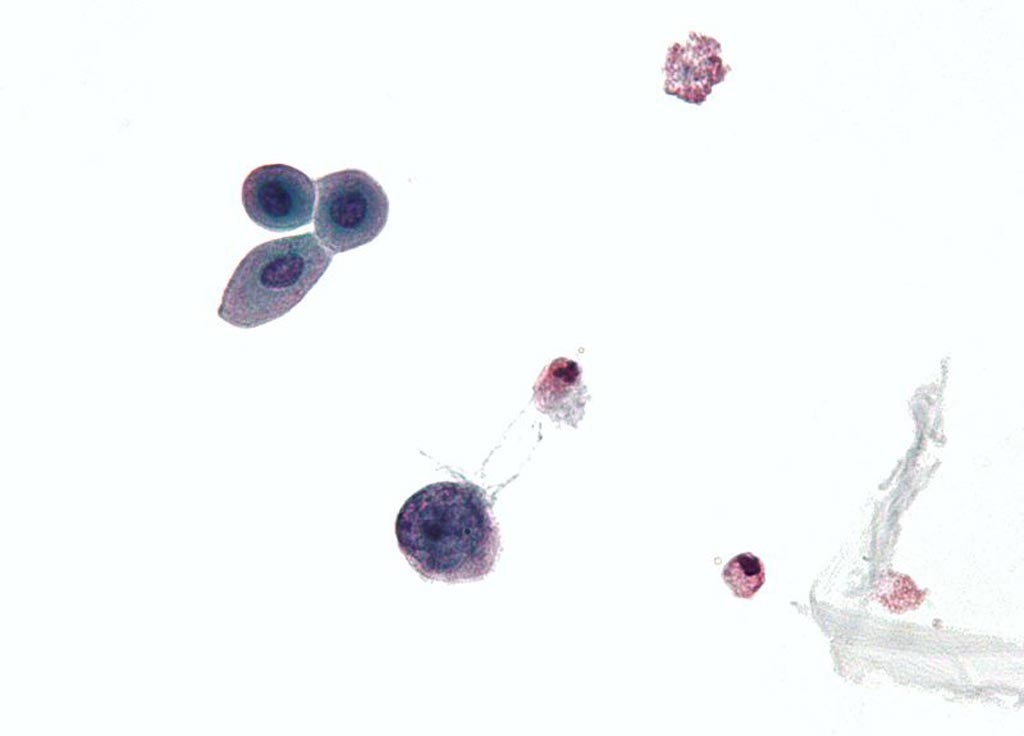
Image: A micrograph of a urine cytology specimen showing a polyomavirus infected cell (Photo courtesy of Wikimedia Commons).
To provide more information about the host-pathogen relationship, investigators at Cornell University (Ithaca, NY, USA) developed a liquid biopsy technique to map cell-free DNA (cfDNA) from clinical samples such as blood and urine. This assay simultaneously quantified the abundance of a large range of viral and bacterial pathogens as well as the degree of host tissue injury from host–microorganism interaction. This was accomplished with a genome-wide measurement of cell-free DNA methylation marks via bisulfite sequencing, a process in which the cell-free DNA was treated with salt to reveal methylation marks.
Bisulfite sequencing to determine the pattern of methylation was performed following treatment of DNA with bisulfite. Treatment of DNA with bisulfite converted cytosine residues to uracil, but left 5-methylcytosine residues unaffected. Therefore, DNA that had been treated with bisulfite retained only methylated cytosines. Bisulfite treatment introduced specific changes in the DNA sequence that depended on the methylation status of individual cytosine residues, yielding single-nucleotide resolution information about the methylation status of a segment of DNA.
The investigators applied the new assay technique to analyze 51 urinary cfDNA isolates collected from a cohort of kidney transplant recipients with and without bacterial and viral infection of the urinary tract. They found that the cell and tissue types of origin of urinary cfDNA could be derived from its genome-wide profile of methylation marks, and strongly depended on infection status. Furthermore, they found evidence of kidney and bladder tissue damage due to viral and bacterial infection, respectively, and of the recruitment of neutrophils to the urinary tract during infection. Through direct comparison to conventional DNA sequencing as well as clinical tests of infection, they found that this assay accurately captured the bacterial and viral composition of the sample.
In particular, the investigators demonstrated that kidney-specific urine cell-free DNA was higher in individuals with BK polyomavirus (BKV nephropathy) as compared to those with BKV replication alone and those with no BKV replication, suggesting a role for this assay to monitor kidney damage in the face of active viral replication and infection.
Senior author Dr. Iwijn De Vlaminck, professor of biomedical engineering at Cornell University, said, "So we developed an assay that would simultaneously inform us about the presence or absence of a wide range of pathogens, but at the same time would also tell us about the injury of different host tissues. The combined information enables us to more definitively say whether a person is dealing with disease or not. But there was still a big gap to assess whether that organism is actually causing disease. That is really a critical question, because some organisms are just commensals, they live side by side with the host. Our guts are filled with microbes, but those microbes may not be the reason you are suffering from disease. In a way, you are infected. You are colonized, but that is just part of normal biology."
The new assay method was described in the August 26, 2019, online edition of the journal Proceedings of the [U.S.] National Academy of Sciences.
Related Links:
Cornell University