New Tool Developed for Diagnosis of Chronic HBV Infection
|
By LabMedica International staff writers Posted on 23 Oct 2019 |
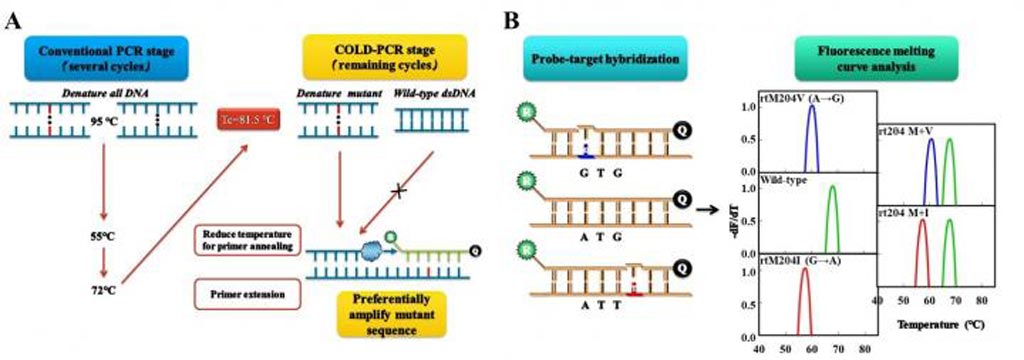
Image: Diagram workflow for co-amplification at lower denaturation temperature PCR (COLD-PCR)/fluorescence melting curve analysis (FMCA) (Photo courtesy of The First Affiliated Hospital of Fujian Medical University).
A team of Chinese researchers has described the development of a new and powerful tool for the diagnosis and treatment of chronic hepatitis B virus (HBV) infection.
Methods currently used to diagnose and monitor chronic hepatitis B (CHB) infection are for the most part based on dynamic and real-time HBV DNA, genotype, and reverse transcriptase (RT) mutation analysis. However, the methods used to perform these analyses are limited by poor sensitivity or inability to detect more than one mutation at a time. Other methods are too cumbersome or expensive for clinical use.
Investigators at Fujian Medical University (Peoples Republic of China) have improved this situation by establishing a highly sensitive co-amplification at lower denaturation temperature PCR (COLD-PCR) coupled with probe-based fluorescence melting curve analysis (FMCA) for precision diagnosis of CHB patients.
COLD-PCR (co-amplification at lower denaturation temperature-PCR) is a modified Polymerase Chain Reaction (PCR) protocol that enriches variant alleles from a mixture of wildtype and mutation-containing DNA. The ability to preferentially amplify and identify minority alleles and low-level somatic DNA mutations in the presence of excess wildtype alleles is useful for the detection of mutations.
Melting curve analysis is an assessment of the dissociation characteristics of double-stranded DNA during heating. As the temperature is raised, the double strand begins to dissociate leading to a rise in the absorbance intensity, hyperchromicity. The temperature at which 50% of DNA is denatured is known as the melting temperature. The information gathered can be used to infer the presence and identity of single-nucleotide polymorphism (SNP) mutations.
The COLD-PCR/FMCA method was shown to be able to detect HBV DNA, genotypes, and RT mutations, simultaneously. The analytical performance of this method, including imprecision, accuracy, sensitivity, detection limits, linear range, and its ability to detect minor variants was systematically evaluated.
Results revealed that the COLD-PCR/FMCA method could detect HBV mutations at much lower concentrations than other techniques such as PCR/FMCA or PCR Sanger sequencing (1% vs. 10% vs. 20%, respectively). The new technique could also distinguish different phases of HBV infection according to the proportion and type of mutations as well as by detecting HBV DNA.
"Guidelines have confirmed that dynamic monitoring of HBV DNA, genotypes, and reverse transcriptase (RT) mutant DNA is of great importance to assess infection status, predict disease progression, and judge treatment efficacy in HBV-infected patients," senior author Dr. Qishui Ou, a researcher in laboratory medicine at The First Afilliated Hospital of Fujian Medical University. "We believe COLD-PCR/FMCA provides a powerful laboratory tool for precise diagnosis and treatment of HBV-infected patients."
"Our goal was to establish a more practical and inexpensive method with high sensitivity to detect genotype and RT mutations while detecting HBV DNA," said Dr. Ou. "Until now there have not been high-throughput approaches to detect HBV DNA, genotype, and RT mutations simultaneously. Therefore, it is necessary to establish a more practical and inexpensive method with high sensitivity to detect genotype and RT mutations while detecting HBV DNA. COLD-PCR/FMCA has that potential."
The study was published in the October 10, 2019, online edition of the Journal of Molecular Diagnostics.
Related Links:
Fujian Medical University
Methods currently used to diagnose and monitor chronic hepatitis B (CHB) infection are for the most part based on dynamic and real-time HBV DNA, genotype, and reverse transcriptase (RT) mutation analysis. However, the methods used to perform these analyses are limited by poor sensitivity or inability to detect more than one mutation at a time. Other methods are too cumbersome or expensive for clinical use.
Investigators at Fujian Medical University (Peoples Republic of China) have improved this situation by establishing a highly sensitive co-amplification at lower denaturation temperature PCR (COLD-PCR) coupled with probe-based fluorescence melting curve analysis (FMCA) for precision diagnosis of CHB patients.
COLD-PCR (co-amplification at lower denaturation temperature-PCR) is a modified Polymerase Chain Reaction (PCR) protocol that enriches variant alleles from a mixture of wildtype and mutation-containing DNA. The ability to preferentially amplify and identify minority alleles and low-level somatic DNA mutations in the presence of excess wildtype alleles is useful for the detection of mutations.
Melting curve analysis is an assessment of the dissociation characteristics of double-stranded DNA during heating. As the temperature is raised, the double strand begins to dissociate leading to a rise in the absorbance intensity, hyperchromicity. The temperature at which 50% of DNA is denatured is known as the melting temperature. The information gathered can be used to infer the presence and identity of single-nucleotide polymorphism (SNP) mutations.
The COLD-PCR/FMCA method was shown to be able to detect HBV DNA, genotypes, and RT mutations, simultaneously. The analytical performance of this method, including imprecision, accuracy, sensitivity, detection limits, linear range, and its ability to detect minor variants was systematically evaluated.
Results revealed that the COLD-PCR/FMCA method could detect HBV mutations at much lower concentrations than other techniques such as PCR/FMCA or PCR Sanger sequencing (1% vs. 10% vs. 20%, respectively). The new technique could also distinguish different phases of HBV infection according to the proportion and type of mutations as well as by detecting HBV DNA.
"Guidelines have confirmed that dynamic monitoring of HBV DNA, genotypes, and reverse transcriptase (RT) mutant DNA is of great importance to assess infection status, predict disease progression, and judge treatment efficacy in HBV-infected patients," senior author Dr. Qishui Ou, a researcher in laboratory medicine at The First Afilliated Hospital of Fujian Medical University. "We believe COLD-PCR/FMCA provides a powerful laboratory tool for precise diagnosis and treatment of HBV-infected patients."
"Our goal was to establish a more practical and inexpensive method with high sensitivity to detect genotype and RT mutations while detecting HBV DNA," said Dr. Ou. "Until now there have not been high-throughput approaches to detect HBV DNA, genotype, and RT mutations simultaneously. Therefore, it is necessary to establish a more practical and inexpensive method with high sensitivity to detect genotype and RT mutations while detecting HBV DNA. COLD-PCR/FMCA has that potential."
The study was published in the October 10, 2019, online edition of the Journal of Molecular Diagnostics.
Related Links:
Fujian Medical University
Latest BioResearch News
- Genome Analysis Predicts Likelihood of Neurodisability in Oxygen-Deprived Newborns
- Gene Panel Predicts Disease Progession for Patients with B-cell Lymphoma
- New Method Simplifies Preparation of Tumor Genomic DNA Libraries
- Panel of Genetic Loci Accurately Predicts Risk of Developing Gout
- Disrupted TGFB Signaling Linked to Increased Cancer-Related Bacteria
- Gene Fusion Protein Proposed as Prostate Cancer Biomarker
- NIV Test to Diagnose and Monitor Vascular Complications in Diabetes
- Semen Exosome MicroRNA Proves Biomarker for Prostate Cancer
- Genetic Loci Link Plasma Lipid Levels to CVD Risk
- Newly Identified Gene Network Aids in Early Diagnosis of Autism Spectrum Disorder
- Link Confirmed between Living in Poverty and Developing Diseases
- Genomic Study Identifies Kidney Disease Loci in Type I Diabetes Patients
- Liquid Biopsy More Effective for Analyzing Tumor Drug Resistance Mutations
- New Liquid Biopsy Assay Reveals Host-Pathogen Interactions
- Method Developed for Enriching Trophoblast Population in Samples
- RNA-Based Test Developed for HPV Detection and Cancer Diagnosis
Channels
Clinical Chemistry
view channel
Electronic Nose Smells Early Signs of Ovarian Cancer in Blood
Ovarian cancer is often diagnosed at a late stage because its symptoms are vague and resemble those of more common conditions. Unlike breast cancer, there is currently no reliable screening method, and... Read more
Simple Blood Test Offers New Path to Alzheimer’s Assessment in Primary Care
Timely evaluation of cognitive symptoms in primary care is often limited by restricted access to specialized diagnostics and invasive confirmatory procedures. Clinicians need accessible tools to determine... Read moreMolecular Diagnostics
view channel
New Blood Test Score Detects Hidden Alcohol-Related Liver Disease
Fatty liver disease affects nearly one in three adults worldwide and can be driven by metabolic conditions such as obesity and diabetes or by excessive alcohol use. In routine care, it is often difficult... Read more
New Blood Test Predicts Who Will Most Likely Live Longer
As people age, it becomes increasingly difficult to determine who is likely to maintain stable health and who may face serious decline. Traditional indicators such as age, cholesterol, and physical activity... Read moreHematology
view channel
Rapid Cartridge-Based Test Aims to Expand Access to Hemoglobin Disorder Diagnosis
Sickle cell disease and beta thalassemia are hemoglobin disorders that often require referral to specialized laboratories for definitive diagnosis, delaying results for patients and clinicians.... Read more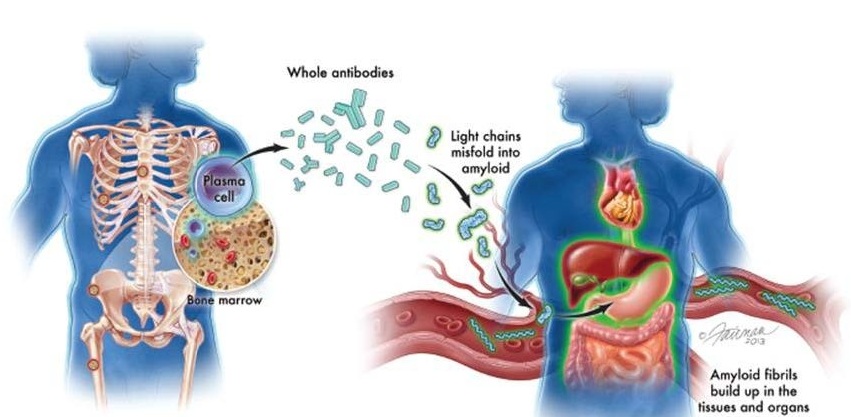
New Guidelines Aim to Improve AL Amyloidosis Diagnosis
Light chain (AL) amyloidosis is a rare, life-threatening bone marrow disorder in which abnormal amyloid proteins accumulate in organs. Approximately 3,260 people in the United States are diagnosed... Read moreImmunology
view channel
New Biomarker Predicts Chemotherapy Response in Triple-Negative Breast Cancer
Triple-negative breast cancer is an aggressive form of breast cancer in which patients often show widely varying responses to chemotherapy. Predicting who will benefit from treatment remains challenging,... Read moreBlood Test Identifies Lung Cancer Patients Who Can Benefit from Immunotherapy Drug
Small cell lung cancer (SCLC) is an aggressive disease with limited treatment options, and even newly approved immunotherapies do not benefit all patients. While immunotherapy can extend survival for some,... Read more
Whole-Genome Sequencing Approach Identifies Cancer Patients Benefitting From PARP-Inhibitor Treatment
Targeted cancer therapies such as PARP inhibitors can be highly effective, but only for patients whose tumors carry specific DNA repair defects. Identifying these patients accurately remains challenging,... Read more
Ultrasensitive Liquid Biopsy Demonstrates Efficacy in Predicting Immunotherapy Response
Immunotherapy has transformed cancer treatment, but only a small proportion of patients experience lasting benefit, with response rates often remaining between 10% and 20%. Clinicians currently lack reliable... Read moreMicrobiology
view channel
Hidden Gut Viruses Linked to Colorectal Cancer Risk
Colorectal cancer (CRC) remains a leading cause of cancer mortality in many Western countries, and existing risk-stratification approaches leave substantial room for improvement. Although age, diet, and... Read more
Three-Test Panel Launched for Detection of Liver Fluke Infections
Parasitic liver fluke infections remain endemic in parts of Asia, where transmission commonly occurs through consumption of raw freshwater fish or aquatic plants. Chronic infection is a well-established... Read morePathology
view channel
Urine Specimen Collection System Improves Diagnostic Accuracy and Efficiency
Urine testing is a critical, non-invasive diagnostic tool used to detect conditions such as pregnancy, urinary tract infections, metabolic disorders, cancer, and kidney disease. However, contaminated or... Read more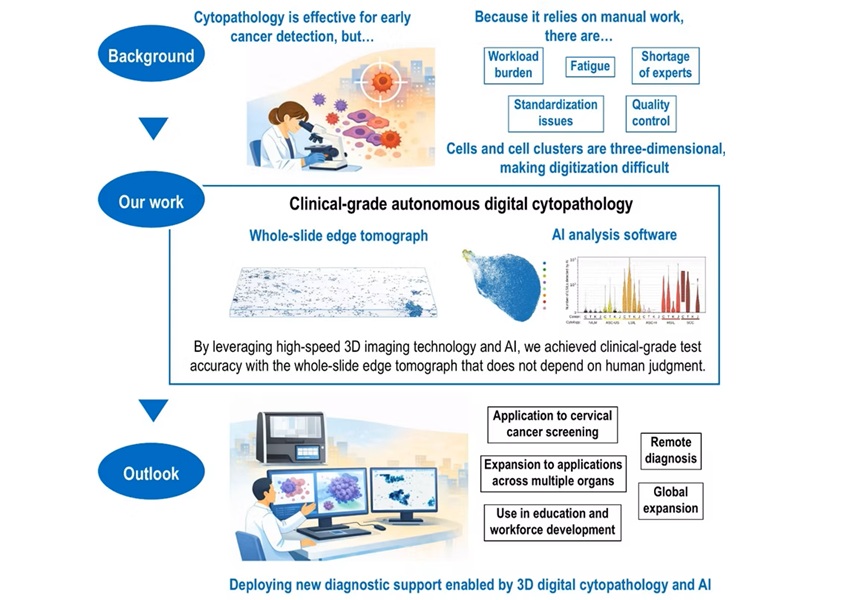
AI-Powered 3D Scanning System Speeds Cancer Screening
Cytology remains a cornerstone of cancer detection, requiring specialists to examine bodily fluids and cells under a microscope. This labor-intensive process involves inspecting up to one million cells... Read moreTechnology
view channel
Blood Test “Clocks” Predict Start of Alzheimer’s Symptoms
More than 7 million Americans live with Alzheimer’s disease, and related health and long-term care costs are projected to reach nearly USD 400 billion in 2025. The disease has no cure, and symptoms often... Read more
AI-Powered Biomarker Predicts Liver Cancer Risk
Liver cancer, or hepatocellular carcinoma, causes more than 800,000 deaths worldwide each year and often goes undetected until late stages. Even after treatment, recurrence rates reach 70% to 80%, contributing... Read more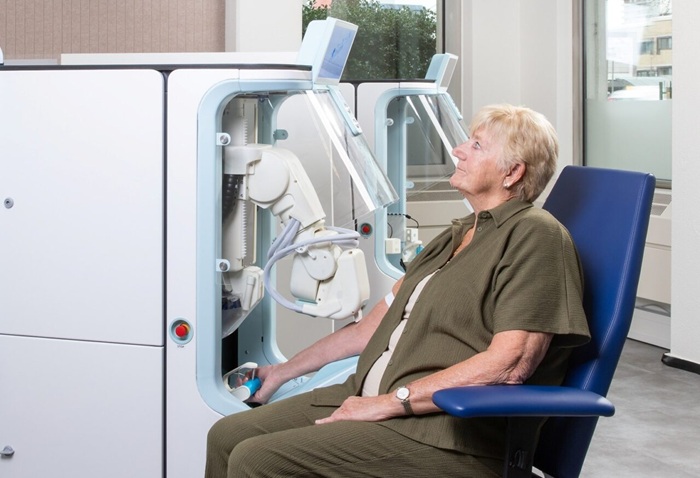
Robotic Technology Unveiled for Automated Diagnostic Blood Draws
Routine diagnostic blood collection is a high‑volume task that can strain staffing and introduce human‑dependent variability, with downstream implications for sample quality and patient experience.... Read more
ADLM Launches First-of-Its-Kind Data Science Program for Laboratory Medicine Professionals
Clinical laboratories generate billions of test results each year, creating a treasure trove of data with the potential to support more personalized testing, improve operational efficiency, and enhance patient care.... Read moreIndustry
view channel
Cepheid Joins CDC Initiative to Strengthen U.S. Pandemic Testing Preparednesss
Cepheid (Sunnyvale, CA, USA) has been selected by the U.S. Centers for Disease Control and Prevention (CDC) as one of four national collaborators in a federal initiative to speed rapid diagnostic technologies... Read more
QuidelOrtho Collaborates with Lifotronic to Expand Global Immunoassay Portfolio
QuidelOrtho (San Diego, CA, USA) has entered a long-term strategic supply agreement with Lifotronic Technology (Shenzhen, China) to expand its global immunoassay portfolio and accelerate customer access... Read more