New Method Simplifies Preparation of Tumor Genomic DNA Libraries
|
By LabMedica International staff writers Posted on 28 Oct 2019 |

Image: A diagram of construction of a genomic library (Photo courtesy of Wikimedia Commons).
A method has been described that simplifies preparation of tumor genomic DNA libraries by employing restriction enzymes and in vitro transcription to barcode and amplify genomic DNA prior to library construction.
In general, a genomic library is the collection of the total genomic DNA from a single organism. The DNA is stored in a population of identical vectors, each containing a different insert of DNA. In order to construct a genomic library, the organism's DNA is extracted from cells and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. The fragments are then inserted into the vector using DNA ligase.
Copy number alterations or variations are common features of cancer cells. Within the same tumor, cells belonging to different anatomical areas of the tumor may carry different variations. Tumors with many variations are typically very aggressive and tend to be resistant to treatment.
Current strategies for massively parallel sequencing of tumor genomic DNA mainly rely on library indexing in the final steps of library preparation. This procedure is costly and time-consuming, since a library must be generated separately for each sample. Furthermore, whole-genome amplification requires intact DNA and thus is problematic in fixed tissue samples, in particular formalin-fixed, paraffin-embedded (FFPE) specimens, which still represent a cornerstone in pathology.
To overcome these limitations, investigators at the Karolinska Institutet (Solna, Sweden) developed a method, which they named CUTseq, that combined restriction endonucleases with in vitro transcription (IVT) to construct highly multiplexed DNA libraries for reduced representation genome sequencing of multiple samples in parallel.
The investigators showed that CUTseq could be used to barcode gDNA extracted from both non-fixed and fixed samples, including old archival FFPE tissue sections. They benchmarked CUTseq by comparing it with a widely used method of DNA library preparation and demonstrated that CUTseq could be used for reduced representation genome and exome sequencing, enabling reproducible DNA copy number profiling and single-nucleotide variant (SNV) calling in both cell and low-input FFPE tissue samples.
The investigators demonstrated an application of CUTseq for assessing genetic differences within tumors by profiling DNA copy number levels in multiple small regions of individual FFPE tumor sections. Finally, they described a workflow for rapid and cost-effective preparation of highly multiplexed CUTseq libraries, which could be applied in the context of high-throughput genetic screens and for cell line authentication.
"I expect that CUTseq will find many useful applications in cancer diagnostics," said senior author Dr. Nicola Crosetto, senior researcher in medical biochemistry and biophysics at the Karolinska Institutet. "Multi-region tumor sequencing is going to be increasingly used in the diagnostic setting, in order to identify patients with highly heterogeneous tumors that need to be treated more aggressively. I believe that our method can play a leading role here."
The study was published in the October 18, 2019, online edition of the journal Nature Communications.
Related Links:
Karolinska Institutet
In general, a genomic library is the collection of the total genomic DNA from a single organism. The DNA is stored in a population of identical vectors, each containing a different insert of DNA. In order to construct a genomic library, the organism's DNA is extracted from cells and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. The fragments are then inserted into the vector using DNA ligase.
Copy number alterations or variations are common features of cancer cells. Within the same tumor, cells belonging to different anatomical areas of the tumor may carry different variations. Tumors with many variations are typically very aggressive and tend to be resistant to treatment.
Current strategies for massively parallel sequencing of tumor genomic DNA mainly rely on library indexing in the final steps of library preparation. This procedure is costly and time-consuming, since a library must be generated separately for each sample. Furthermore, whole-genome amplification requires intact DNA and thus is problematic in fixed tissue samples, in particular formalin-fixed, paraffin-embedded (FFPE) specimens, which still represent a cornerstone in pathology.
To overcome these limitations, investigators at the Karolinska Institutet (Solna, Sweden) developed a method, which they named CUTseq, that combined restriction endonucleases with in vitro transcription (IVT) to construct highly multiplexed DNA libraries for reduced representation genome sequencing of multiple samples in parallel.
The investigators showed that CUTseq could be used to barcode gDNA extracted from both non-fixed and fixed samples, including old archival FFPE tissue sections. They benchmarked CUTseq by comparing it with a widely used method of DNA library preparation and demonstrated that CUTseq could be used for reduced representation genome and exome sequencing, enabling reproducible DNA copy number profiling and single-nucleotide variant (SNV) calling in both cell and low-input FFPE tissue samples.
The investigators demonstrated an application of CUTseq for assessing genetic differences within tumors by profiling DNA copy number levels in multiple small regions of individual FFPE tumor sections. Finally, they described a workflow for rapid and cost-effective preparation of highly multiplexed CUTseq libraries, which could be applied in the context of high-throughput genetic screens and for cell line authentication.
"I expect that CUTseq will find many useful applications in cancer diagnostics," said senior author Dr. Nicola Crosetto, senior researcher in medical biochemistry and biophysics at the Karolinska Institutet. "Multi-region tumor sequencing is going to be increasingly used in the diagnostic setting, in order to identify patients with highly heterogeneous tumors that need to be treated more aggressively. I believe that our method can play a leading role here."
The study was published in the October 18, 2019, online edition of the journal Nature Communications.
Related Links:
Karolinska Institutet
Latest BioResearch News
- Genome Analysis Predicts Likelihood of Neurodisability in Oxygen-Deprived Newborns
- Gene Panel Predicts Disease Progession for Patients with B-cell Lymphoma
- New Tool Developed for Diagnosis of Chronic HBV Infection
- Panel of Genetic Loci Accurately Predicts Risk of Developing Gout
- Disrupted TGFB Signaling Linked to Increased Cancer-Related Bacteria
- Gene Fusion Protein Proposed as Prostate Cancer Biomarker
- NIV Test to Diagnose and Monitor Vascular Complications in Diabetes
- Semen Exosome MicroRNA Proves Biomarker for Prostate Cancer
- Genetic Loci Link Plasma Lipid Levels to CVD Risk
- Newly Identified Gene Network Aids in Early Diagnosis of Autism Spectrum Disorder
- Link Confirmed between Living in Poverty and Developing Diseases
- Genomic Study Identifies Kidney Disease Loci in Type I Diabetes Patients
- Liquid Biopsy More Effective for Analyzing Tumor Drug Resistance Mutations
- New Liquid Biopsy Assay Reveals Host-Pathogen Interactions
- Method Developed for Enriching Trophoblast Population in Samples
- RNA-Based Test Developed for HPV Detection and Cancer Diagnosis
Channels
Clinical Chemistry
view channel
New PSA-Based Prognostic Model Improves Prostate Cancer Risk Assessment
Prostate cancer is the second-leading cause of cancer death among American men, and about one in eight will be diagnosed in their lifetime. Screening relies on blood levels of prostate-specific antigen... Read more
Extracellular Vesicles Linked to Heart Failure Risk in CKD Patients
Chronic kidney disease (CKD) affects more than 1 in 7 Americans and is strongly associated with cardiovascular complications, which account for more than half of deaths among people with CKD.... Read moreMolecular Diagnostics
view channel
Diagnostic Device Predicts Treatment Response for Brain Tumors Via Blood Test
Glioblastoma is one of the deadliest forms of brain cancer, largely because doctors have no reliable way to determine whether treatments are working in real time. Assessing therapeutic response currently... Read more
Blood Test Detects Early-Stage Cancers by Measuring Epigenetic Instability
Early-stage cancers are notoriously difficult to detect because molecular changes are subtle and often missed by existing screening tools. Many liquid biopsies rely on measuring absolute DNA methylation... Read more
“Lab-On-A-Disc” Device Paves Way for More Automated Liquid Biopsies
Extracellular vesicles (EVs) are tiny particles released by cells into the bloodstream that carry molecular information about a cell’s condition, including whether it is cancerous. However, EVs are highly... Read more
Blood Test Identifies Inflammatory Breast Cancer Patients at Increased Risk of Brain Metastasis
Brain metastasis is a frequent and devastating complication in patients with inflammatory breast cancer, an aggressive subtype with limited treatment options. Despite its high incidence, the biological... Read moreHematology
view channel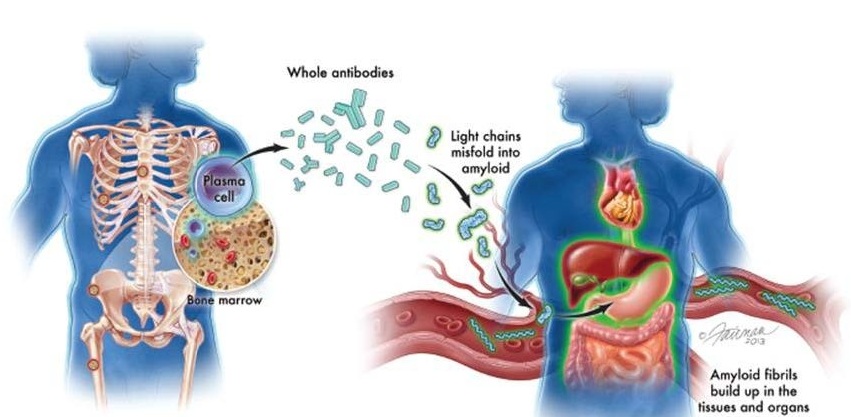
New Guidelines Aim to Improve AL Amyloidosis Diagnosis
Light chain (AL) amyloidosis is a rare, life-threatening bone marrow disorder in which abnormal amyloid proteins accumulate in organs. Approximately 3,260 people in the United States are diagnosed... Read more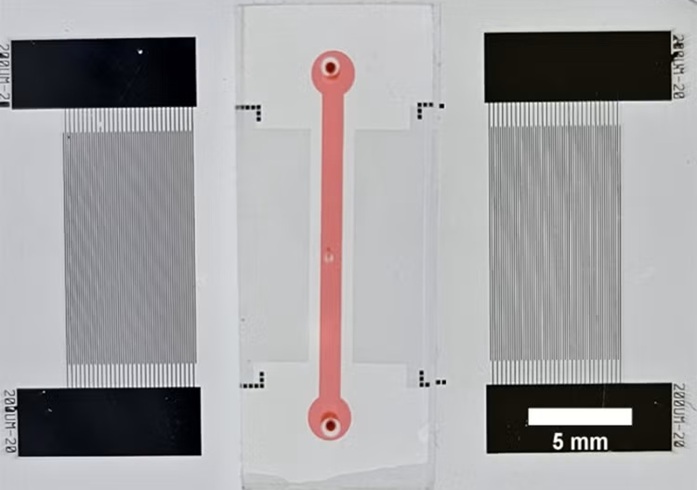
Fast and Easy Test Could Revolutionize Blood Transfusions
Blood transfusions are a cornerstone of modern medicine, yet red blood cells can deteriorate quietly while sitting in cold storage for weeks. Although blood units have a fixed expiration date, cells from... Read more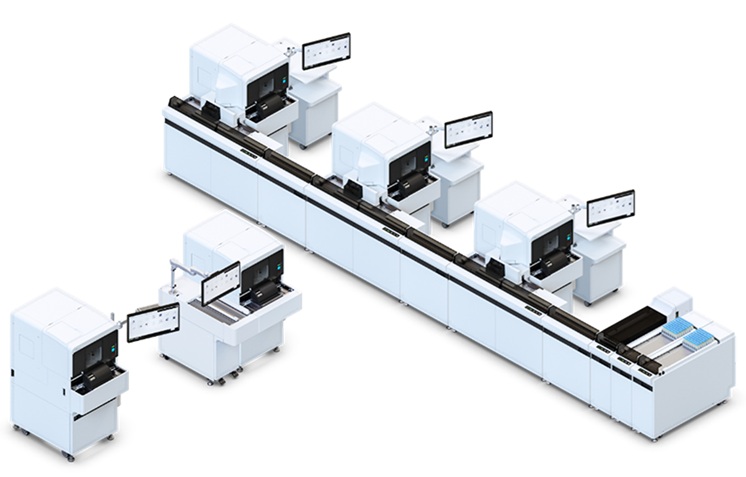
Automated Hemostasis System Helps Labs of All Sizes Optimize Workflow
High-volume hemostasis sections must sustain rapid turnaround while managing reruns and reflex testing. Manual tube handling and preanalytical checks can strain staff time and increase opportunities for error.... Read more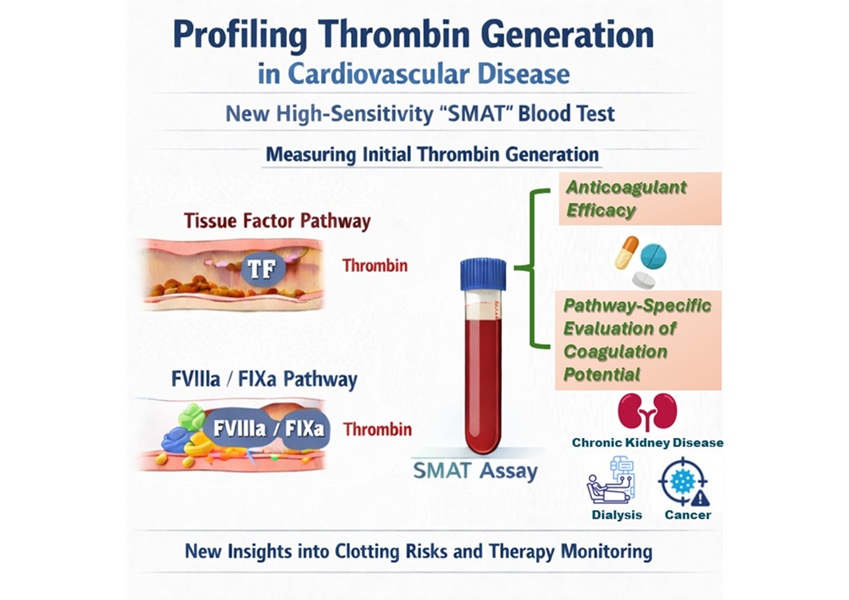
High-Sensitivity Blood Test Improves Assessment of Clotting Risk in Heart Disease Patients
Blood clotting is essential for preventing bleeding, but even small imbalances can lead to serious conditions such as thrombosis or dangerous hemorrhage. In cardiovascular disease, clinicians often struggle... Read moreImmunology
view channelBlood Test Identifies Lung Cancer Patients Who Can Benefit from Immunotherapy Drug
Small cell lung cancer (SCLC) is an aggressive disease with limited treatment options, and even newly approved immunotherapies do not benefit all patients. While immunotherapy can extend survival for some,... Read more
Whole-Genome Sequencing Approach Identifies Cancer Patients Benefitting From PARP-Inhibitor Treatment
Targeted cancer therapies such as PARP inhibitors can be highly effective, but only for patients whose tumors carry specific DNA repair defects. Identifying these patients accurately remains challenging,... Read more
Ultrasensitive Liquid Biopsy Demonstrates Efficacy in Predicting Immunotherapy Response
Immunotherapy has transformed cancer treatment, but only a small proportion of patients experience lasting benefit, with response rates often remaining between 10% and 20%. Clinicians currently lack reliable... Read moreMicrobiology
view channel
Comprehensive Review Identifies Gut Microbiome Signatures Associated With Alzheimer’s Disease
Alzheimer’s disease affects approximately 6.7 million people in the United States and nearly 50 million worldwide, yet early cognitive decline remains difficult to characterize. Increasing evidence suggests... Read moreAI-Powered Platform Enables Rapid Detection of Drug-Resistant C. Auris Pathogens
Infections caused by the pathogenic yeast Candida auris pose a significant threat to hospitalized patients, particularly those with weakened immune systems or those who have invasive medical devices.... Read morePathology
view channel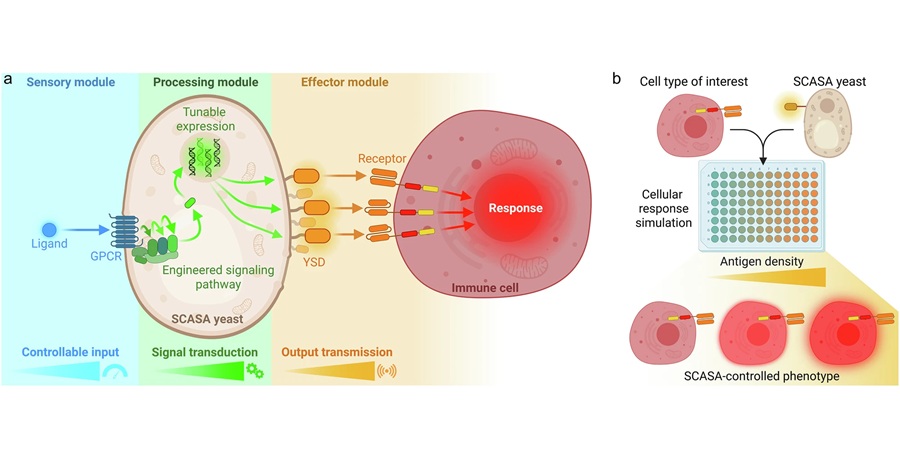
Engineered Yeast Cells Enable Rapid Testing of Cancer Immunotherapy
Developing new cancer immunotherapies is a slow, costly, and high-risk process, particularly for CAR T cell treatments that must precisely recognize cancer-specific antigens. Small differences in tumor... Read more
First-Of-Its-Kind Test Identifies Autism Risk at Birth
Autism spectrum disorder is treatable, and extensive research shows that early intervention can significantly improve cognitive, social, and behavioral outcomes. Yet in the United States, the average age... Read moreTechnology
view channel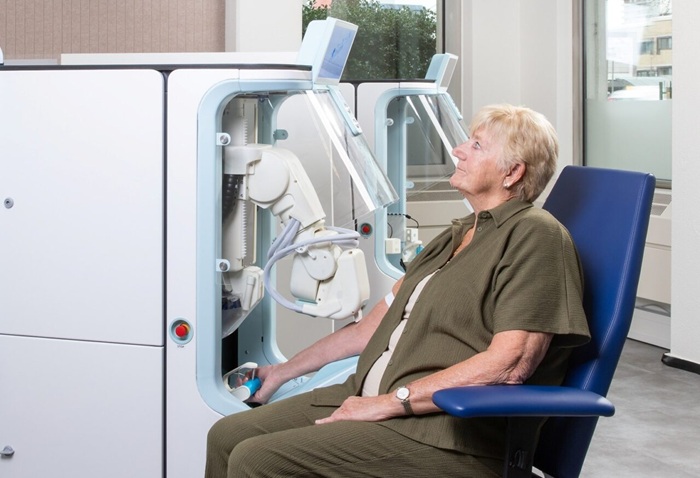
Robotic Technology Unveiled for Automated Diagnostic Blood Draws
Routine diagnostic blood collection is a high‑volume task that can strain staffing and introduce human‑dependent variability, with downstream implications for sample quality and patient experience.... Read more
ADLM Launches First-of-Its-Kind Data Science Program for Laboratory Medicine Professionals
Clinical laboratories generate billions of test results each year, creating a treasure trove of data with the potential to support more personalized testing, improve operational efficiency, and enhance patient care.... Read moreAptamer Biosensor Technology to Transform Virus Detection
Rapid and reliable virus detection is essential for controlling outbreaks, from seasonal influenza to global pandemics such as COVID-19. Conventional diagnostic methods, including cell culture, antigen... Read more
AI Models Could Predict Pre-Eclampsia and Anemia Earlier Using Routine Blood Tests
Pre-eclampsia and anemia are major contributors to maternal and child mortality worldwide, together accounting for more than half a million deaths each year and leaving millions with long-term health complications.... Read moreIndustry
view channelNew Collaboration Brings Automated Mass Spectrometry to Routine Laboratory Testing
Mass spectrometry is a powerful analytical technique that identifies and quantifies molecules based on their mass and electrical charge. Its high selectivity, sensitivity, and accuracy make it indispensable... Read more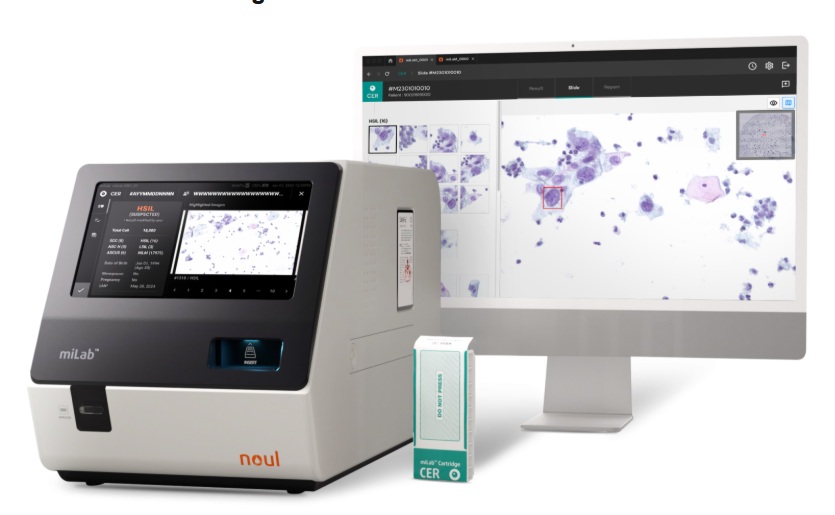
AI-Powered Cervical Cancer Test Set for Major Rollout in Latin America
Noul Co., a Korean company specializing in AI-based blood and cancer diagnostics, announced it will supply its intelligence (AI)-based miLab CER cervical cancer diagnostic solution to Mexico under a multi‑year... Read more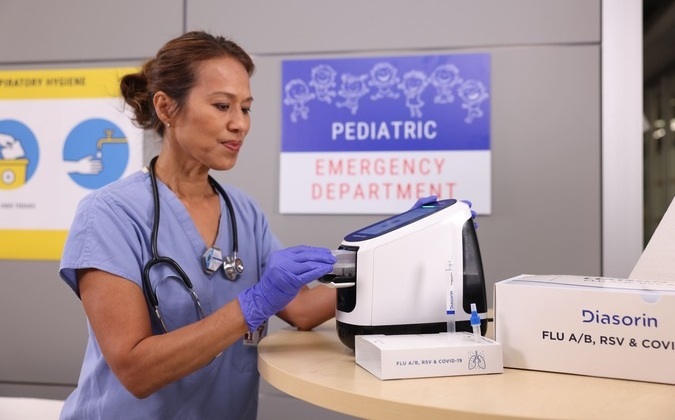
Diasorin and Fisher Scientific Enter into US Distribution Agreement for Molecular POC Platform
Diasorin (Saluggia, Italy) has entered into an exclusive distribution agreement with Fisher Scientific, part of Thermo Fisher Scientific (Waltham, MA, USA), for the LIAISON NES molecular point-of-care... Read more