A Tool to Predict Tuberculosis Drug Resistance Now Available Online
|
By LabMedica International staff writers Posted on 09 Jun 2015 |
![Image: Mycobacterium tuberculosis (stained purple) in a tissue specimen (blue) (Photo courtesy of the CDC - [US] Centers for Disease Control and Prevention). Image: Mycobacterium tuberculosis (stained purple) in a tissue specimen (blue) (Photo courtesy of the CDC - [US] Centers for Disease Control and Prevention).](https://globetechcdn.com/mobile_labmedica/images/stories/articles/article_images/2015-06-09/GMS-170.jpg)
Image: Mycobacterium tuberculosis (stained purple) in a tissue specimen (blue) (Photo courtesy of the CDC - [US] Centers for Disease Control and Prevention).
A new online tool for the rapid analysis of whole genome sequence data is set to aid clinicians predict whether a particular patient's tuberculosis (TB) will be susceptible to frequently prescribed antibiotics.
The World Health Organization (WHO) estimated that 5% of the world's 11 million tuberculosis patients have multi-drug-resistance disease (MDR-TB) and that 480,000 new cases arose during 2013 alone. Of those approximately 9% have extensively resistant tuberculosis (XDR-TB) where, in addition to resistance to at least both of the major first line drugs (isoniazid and rifampicin), they also have resistance to two classes of second line drugs used to treat MDR-TB (the fluoroquinolones and the injectable drugs, amikacin, kanamycin, or capreomycin).
Current molecular tests examine limited numbers of mutations in Mycobacterium tuberculosis, the organism that causes TB, and although whole genome sequencing could fully characterize drug resistance, the complexity of data obtained by this technology has restricted their clinical application.
To help solve this problem investigators at the London School of Hygiene & Tropical Medicine (United Kingdom) have created an online tool that analyses and interprets genome sequence data to predict resistance to 11 drugs used for the treatment of TB. Initially, a library (1,325 mutations) predictive of drug resistance for 15 anti-tuberculosis drugs was compiled and then validated for 11 of them using genomic-phenotypic data from 792 strains. A rapid online "TB-Profiler" tool was developed to report drug resistance and strain-type profiles directly from raw sequences. The TB-Profiler tool is available on the Internet (Please see Related Links below).
Senior author Dr. Taane Clark, reader in genetic epidemiology and statistical genomics at the London School of Hygiene & Tropical Medicine, said, "Sequencing already assists patient management for a number of conditions such as HIV, but now that it is possible to sequence M. tuberculosis from sputum from suspected multi-drug resistance patients means it has a role in the management of tuberculosis. We have developed a prototype to guide treatment of patients with drug resistant disease, where personalized medicine and treatment offers improved rates of cure."
Complete information regarding the new online tool was published in the May 27, 2015, online edition of the journal Genome Medicine.
Related Links:
TB-Profiler tool
London School of Hygiene & Tropical Medicine
The World Health Organization (WHO) estimated that 5% of the world's 11 million tuberculosis patients have multi-drug-resistance disease (MDR-TB) and that 480,000 new cases arose during 2013 alone. Of those approximately 9% have extensively resistant tuberculosis (XDR-TB) where, in addition to resistance to at least both of the major first line drugs (isoniazid and rifampicin), they also have resistance to two classes of second line drugs used to treat MDR-TB (the fluoroquinolones and the injectable drugs, amikacin, kanamycin, or capreomycin).
Current molecular tests examine limited numbers of mutations in Mycobacterium tuberculosis, the organism that causes TB, and although whole genome sequencing could fully characterize drug resistance, the complexity of data obtained by this technology has restricted their clinical application.
To help solve this problem investigators at the London School of Hygiene & Tropical Medicine (United Kingdom) have created an online tool that analyses and interprets genome sequence data to predict resistance to 11 drugs used for the treatment of TB. Initially, a library (1,325 mutations) predictive of drug resistance for 15 anti-tuberculosis drugs was compiled and then validated for 11 of them using genomic-phenotypic data from 792 strains. A rapid online "TB-Profiler" tool was developed to report drug resistance and strain-type profiles directly from raw sequences. The TB-Profiler tool is available on the Internet (Please see Related Links below).
Senior author Dr. Taane Clark, reader in genetic epidemiology and statistical genomics at the London School of Hygiene & Tropical Medicine, said, "Sequencing already assists patient management for a number of conditions such as HIV, but now that it is possible to sequence M. tuberculosis from sputum from suspected multi-drug resistance patients means it has a role in the management of tuberculosis. We have developed a prototype to guide treatment of patients with drug resistant disease, where personalized medicine and treatment offers improved rates of cure."
Complete information regarding the new online tool was published in the May 27, 2015, online edition of the journal Genome Medicine.
Related Links:
TB-Profiler tool
London School of Hygiene & Tropical Medicine
Latest Microbiology News
- Rapid Sequencing Could Transform Tuberculosis Care
- Blood-Based Viral Signature Identified in Crohn’s Disease
- Hidden Gut Viruses Linked to Colorectal Cancer Risk
- Three-Test Panel Launched for Detection of Liver Fluke Infections
- Rapid Test Promises Faster Answers for Drug-Resistant Infections
- CRISPR-Based Technology Neutralizes Antibiotic-Resistant Bacteria
- Comprehensive Review Identifies Gut Microbiome Signatures Associated With Alzheimer’s Disease
- AI-Powered Platform Enables Rapid Detection of Drug-Resistant C. Auris Pathogens
- New Test Measures How Effectively Antibiotics Kill Bacteria
- New Antimicrobial Stewardship Standards for TB Care to Optimize Diagnostics
- New UTI Diagnosis Method Delivers Antibiotic Resistance Results 24 Hours Earlier
- Breakthroughs in Microbial Analysis to Enhance Disease Prediction
- Blood-Based Diagnostic Method Could Identify Pediatric LRTIs
- Rapid Diagnostic Test Matches Gold Standard for Sepsis Detection
- Rapid POC Tuberculosis Test Provides Results Within 15 Minutes
- Rapid Assay Identifies Bloodstream Infection Pathogens Directly from Patient Samples
Channels
Clinical Chemistry
view channelNew Blood Test Index Offers Earlier Detection of Liver Scarring
Metabolic fatty liver disease is highly prevalent and often silent, yet it can progress to fibrosis, cirrhosis, and liver failure. Current first-line blood test scores frequently return indeterminate results,... Read more
Electronic Nose Smells Early Signs of Ovarian Cancer in Blood
Ovarian cancer is often diagnosed at a late stage because its symptoms are vague and resemble those of more common conditions. Unlike breast cancer, there is currently no reliable screening method, and... Read moreMolecular Diagnostics
view channel
New Test Detects Alzheimer’s by Analyzing Altered Protein Shapes in Blood
Alzheimer’s disease begins developing years before memory loss or other symptoms become visible. Misfolded proteins gradually accumulate in the brain, disrupting normal cellular processes.... Read more
New Diagnostic Markers for Multiple Sclerosis Discovered in Cerebrospinal Fluid
Multiple sclerosis (MS) affects nearly three million people worldwide and can cause symptoms such as numbness, visual disturbances, fatigue, and neurological disability. Diagnosing the disease can be challenging... Read moreHematology
view channel
Rapid Cartridge-Based Test Aims to Expand Access to Hemoglobin Disorder Diagnosis
Sickle cell disease and beta thalassemia are hemoglobin disorders that often require referral to specialized laboratories for definitive diagnosis, delaying results for patients and clinicians.... Read more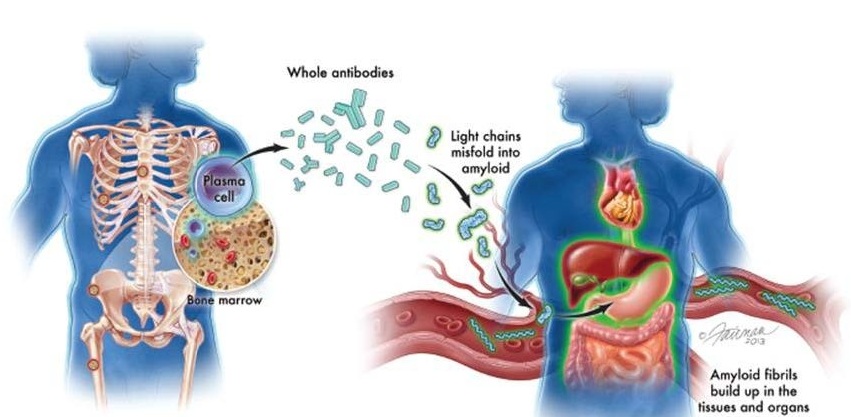
New Guidelines Aim to Improve AL Amyloidosis Diagnosis
Light chain (AL) amyloidosis is a rare, life-threatening bone marrow disorder in which abnormal amyloid proteins accumulate in organs. Approximately 3,260 people in the United States are diagnosed... Read moreImmunology
view channel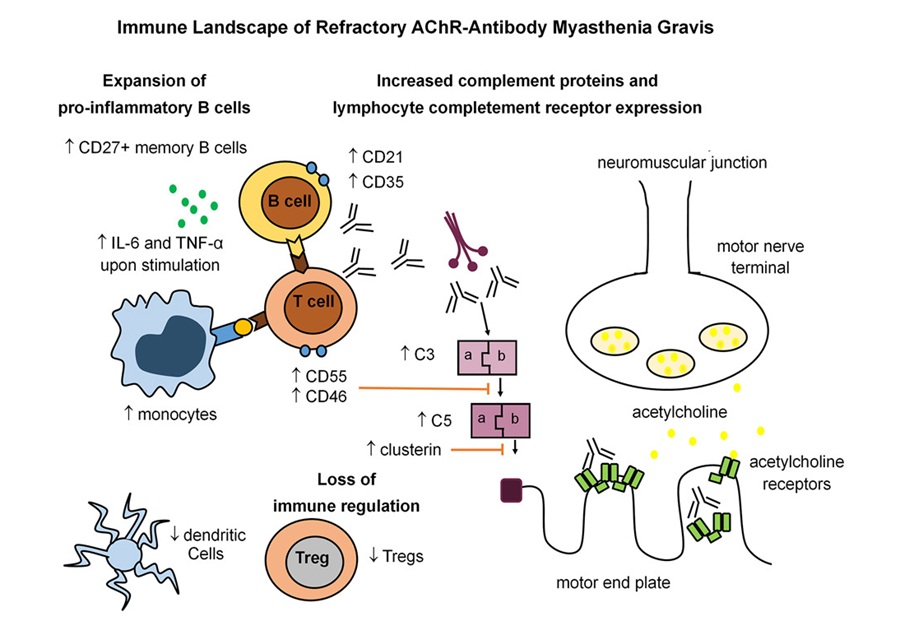
Immune Signature Identified in Treatment-Resistant Myasthenia Gravis
Myasthenia gravis is a rare autoimmune disorder in which immune attack at the neuromuscular junction causes fluctuating weakness that can impair vision, movement, speech, swallowing, and breathing.... Read more
New Biomarker Predicts Chemotherapy Response in Triple-Negative Breast Cancer
Triple-negative breast cancer is an aggressive form of breast cancer in which patients often show widely varying responses to chemotherapy. Predicting who will benefit from treatment remains challenging,... Read moreBlood Test Identifies Lung Cancer Patients Who Can Benefit from Immunotherapy Drug
Small cell lung cancer (SCLC) is an aggressive disease with limited treatment options, and even newly approved immunotherapies do not benefit all patients. While immunotherapy can extend survival for some,... Read more
Whole-Genome Sequencing Approach Identifies Cancer Patients Benefitting From PARP-Inhibitor Treatment
Targeted cancer therapies such as PARP inhibitors can be highly effective, but only for patients whose tumors carry specific DNA repair defects. Identifying these patients accurately remains challenging,... Read morePathology
view channel
Molecular Imaging to Reduce Need for Melanoma Biopsies
Melanoma is the deadliest form of skin cancer and accounts for the vast majority of skin cancer-related deaths. Because early melanomas can closely resemble benign moles, clinicians often rely on visual... Read more
Urine Specimen Collection System Improves Diagnostic Accuracy and Efficiency
Urine testing is a critical, non-invasive diagnostic tool used to detect conditions such as pregnancy, urinary tract infections, metabolic disorders, cancer, and kidney disease. However, contaminated or... Read moreTechnology
view channel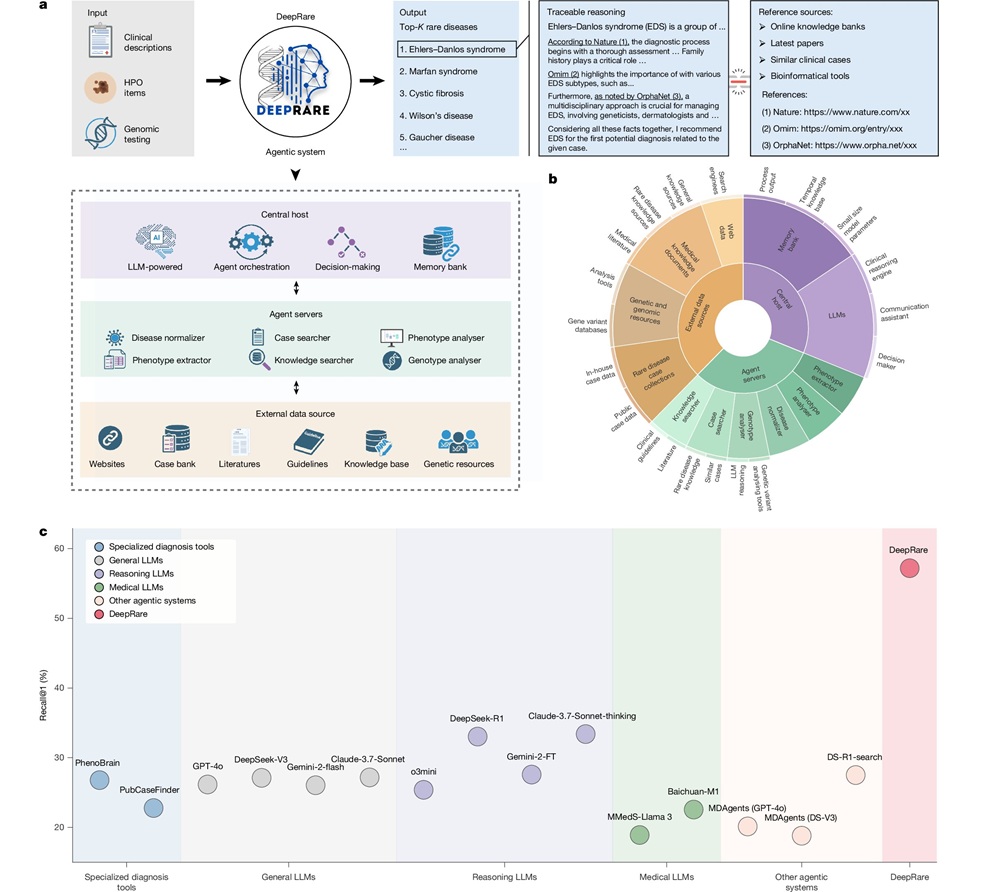
AI Model Outperforms Clinicians in Rare Disease Detection
Rare diseases affect an estimated 300 million people worldwide, yet diagnosis is often protracted and error-prone. Many conditions present with heterogeneous signs that overlap with common disorders, leading... Read more
AI-Driven Diagnostic Demonstrates High Accuracy in Detecting Periprosthetic Joint Infection
Periprosthetic joint infection (PJI) is a rare but serious complication affecting 1% to 2% of primary joint replacement surgeries. The condition occurs when bacteria or fungi infect tissues around an implanted... Read moreIndustry
view channel
Cepheid Joins CDC Initiative to Strengthen U.S. Pandemic Testing Preparednesss
Cepheid (Sunnyvale, CA, USA) has been selected by the U.S. Centers for Disease Control and Prevention (CDC) as one of four national collaborators in a federal initiative to speed rapid diagnostic technologies... Read more
QuidelOrtho Collaborates with Lifotronic to Expand Global Immunoassay Portfolio
QuidelOrtho (San Diego, CA, USA) has entered a long-term strategic supply agreement with Lifotronic Technology (Shenzhen, China) to expand its global immunoassay portfolio and accelerate customer access... Read more