Deep Sequencing of CD34+ Cells Detects Measurable Residual Disease in AML
|
By LabMedica International staff writers Posted on 31 Mar 2022 |

Monitoring of measurable residual disease (MRD) in patients with acute myeloid leukemia is predictive for disease recurrence and may identify patients who benefit from treatment intensification. Current MRD techniques rely on multicolor flow cytometry or molecular methods, but are limited in applicability or sensitivity.
For patients with hematological malignancies such as acute myeloid leukemia (AML) or high risk myelodysplastic syndrome (MDS), the application of allogeneic stem cell transplantation (alloHSCT) often remains the only curative treatment option. Nevertheless, relapse after SCT occurs in 30% to 70% of AML patients and is the major cause of treatment failure, with dismal prognosis and a two-year survival of <20%.
Hematologists at the University Hospital Carl Gustav Carus TU Dresden (Dresden, Germany) and their colleagues retrospectively analyzed 429 peripheral blood (PB) and 55 bone marrow (BM) samples of 40 AML and high-risk MDS patients, with/without molecular relapse based on CD34+ donor chimerism (DC), in complete remission after alloHSCT. The team evaluated the feasibility of a novel approach for MRD detection in PB, which combines immunomagnetic pre-enrichment and fluorescence-activated cell sorting (FACS) for isolation of CD34+ cells with error-reduced targeted next-generation sequencing (NGS).
The team isolated CD34+ / CD117+ cells from mononuclear cells (MNCs) using magnet activated cell sorting (MACS) by positive selection using the CD34+ or the CD117+ Microbead Kit (Miltenyi Biotec, Bergisch-Gladbach, Germany). For FACS-sorting of CD34+ / CD117+ cells, the CD34 or CD117 enriched fractions were incubated with the monoclonal antibodies CD45 FITC / CD34 PE (BD Biosciences, San Jose, CA, USA). Sorting of CD34+ / CD117+ cells was then conducted on a BD FACS Aria II cell sorter, aiming for 5,000-10,000 CD34+ / CD117+ cells and a purity of >90%. DNA extraction For DNA extraction, the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) or the “ZR 168 Viral DNA Kit” (Zymo Research, Orange, CA, USA), for CD34+ / CD117+ cell counts, was used.
The investigators reported that enrichment of CD34+ cells for NGS increased the detection of mutant alleles in PB ~1000-fold (median Variant Allele Frequency [VAF] 1.27% versus 0.0046% in unsorted PB). Although a strong correlation was observed for the parallel analysis of CD34+ PB cells with NGS and DC, the combination of FACS and NGS improved sensitivity for MRD detection in dilution studies ~10-fold to levels of 10-6. In both assays, MRD detection was superior using PB versus BM for CD34+ enrichment. Importantly, NGS on CD34+ PB cells enabled prediction of molecular relapse with high sensitivity (100%) and specificity (91%), and significantly earlier (median 48 days, range 0-281) than by CD34+ DC or NGS of unsorted PB, providing additional time for therapeutic intervention. Moreover, panel sequencing in CD34+ cells allowed the early assessment of clonal trajectories in hematological complete remission.
The authors have proposed a novel, easily accessible and robust method for ultra-sensitive MRD detection in peripheral blood, which is applicable to the vast majority of AML patients. First results demonstrate the feasibility of targeted deep sequencing on CD34+ cells for early relapse prediction in clinical settings, with superior sensitivity and specificity as compared to chimerism-based MRD assessment or the use of unsorted PB for NGS. The study was published on March 23, 2022 in the journal Blood Advances.
Related Links:
University Hospital Carl Gustav Carus TU Dresden
Miltenyi Biotec
BD Biosciences
Qiagen
Zymo Research
Latest Hematology News
- Next Generation Instrument Screens for Hemoglobin Disorders in Newborns
- First 4-in-1 Nucleic Acid Test for Arbovirus Screening to Reduce Risk of Transfusion-Transmitted Infections
- POC Finger-Prick Blood Test Determines Risk of Neutropenic Sepsis in Patients Undergoing Chemotherapy
- First Affordable and Rapid Test for Beta Thalassemia Demonstrates 99% Diagnostic Accuracy
- Handheld White Blood Cell Tracker to Enable Rapid Testing For Infections
- Smart Palm-size Optofluidic Hematology Analyzer Enables POCT of Patients’ Blood Cells
- Automated Hematology Platform Offers High Throughput Analytical Performance
- New Tool Analyzes Blood Platelets Faster, Easily and Accurately
- First Rapid-Result Hematology Analyzer Reports Measures of Infection and Severity at POC
- Bleeding Risk Diagnostic Test to Reduce Preventable Complications in Hospitals
- True POC Hematology Analyzer with Direct Capillary Sampling Enhances Ease-of-Use and Testing Throughput
- Point of Care CBC Analyzer with Direct Capillary Sampling Enhances Ease-of-Use and Testing Throughput
- Blood Test Could Predict Outcomes in Emergency Department and Hospital Admissions
- Novel Technology Diagnoses Immunothrombosis Using Breath Gas Analysis
- Advanced Hematology System Allows Labs to Process Up To 119 Complete Blood Count Results per Hour
- Unique AI-Based Approach Automates Clinical Analysis of Blood Data
Channels
Clinical Chemistry
view channel
3D Printed Point-Of-Care Mass Spectrometer Outperforms State-Of-The-Art Models
Mass spectrometry is a precise technique for identifying the chemical components of a sample and has significant potential for monitoring chronic illness health states, such as measuring hormone levels... Read more.jpg)
POC Biomedical Test Spins Water Droplet Using Sound Waves for Cancer Detection
Exosomes, tiny cellular bioparticles carrying a specific set of proteins, lipids, and genetic materials, play a crucial role in cell communication and hold promise for non-invasive diagnostics.... Read more
Highly Reliable Cell-Based Assay Enables Accurate Diagnosis of Endocrine Diseases
The conventional methods for measuring free cortisol, the body's stress hormone, from blood or saliva are quite demanding and require sample processing. The most common method, therefore, involves collecting... Read moreMolecular Diagnostics
view channelBlood Proteins Could Warn of Cancer Seven Years before Diagnosis
Two studies have identified proteins in the blood that could potentially alert individuals to the presence of cancer more than seven years before the disease is clinically diagnosed. Researchers found... Read moreUltrasound-Aided Blood Testing Detects Cancer Biomarkers from Cells
Ultrasound imaging serves as a noninvasive method to locate and monitor cancerous tumors effectively. However, crucial details about the cancer, such as the specific types of cells and genetic mutations... Read moreImmunology
view channel.jpg)
AI Predicts Tumor-Killing Cells with High Accuracy
Cellular immunotherapy involves extracting immune cells from a patient's tumor, potentially enhancing their cancer-fighting capabilities through engineering, and then expanding and reintroducing them into the body.... Read more
Diagnostic Blood Test for Cellular Rejection after Organ Transplant Could Replace Surgical Biopsies
Transplanted organs constantly face the risk of being rejected by the recipient's immune system which differentiates self from non-self using T cells and B cells. T cells are commonly associated with acute... Read more
AI Tool Precisely Matches Cancer Drugs to Patients Using Information from Each Tumor Cell
Current strategies for matching cancer patients with specific treatments often depend on bulk sequencing of tumor DNA and RNA, which provides an average profile from all cells within a tumor sample.... Read more
Genetic Testing Combined With Personalized Drug Screening On Tumor Samples to Revolutionize Cancer Treatment
Cancer treatment typically adheres to a standard of care—established, statistically validated regimens that are effective for the majority of patients. However, the disease’s inherent variability means... Read moreMicrobiology
view channel
Integrated Solution Ushers New Era of Automated Tuberculosis Testing
Tuberculosis (TB) is responsible for 1.3 million deaths every year, positioning it as one of the top killers globally due to a single infectious agent. In 2022, around 10.6 million people were diagnosed... Read more
Automated Sepsis Test System Enables Rapid Diagnosis for Patients with Severe Bloodstream Infections
Sepsis affects up to 50 million people globally each year, with bacteraemia, formerly known as blood poisoning, being a major cause. In the United States alone, approximately two million individuals are... Read moreEnhanced Rapid Syndromic Molecular Diagnostic Solution Detects Broad Range of Infectious Diseases
GenMark Diagnostics (Carlsbad, CA, USA), a member of the Roche Group (Basel, Switzerland), has rebranded its ePlex® system as the cobas eplex system. This rebranding under the globally renowned cobas name... Read more
Clinical Decision Support Software a Game-Changer in Antimicrobial Resistance Battle
Antimicrobial resistance (AMR) is a serious global public health concern that claims millions of lives every year. It primarily results from the inappropriate and excessive use of antibiotics, which reduces... Read morePathology
view channelHyperspectral Dark-Field Microscopy Enables Rapid and Accurate Identification of Cancerous Tissues
Breast cancer remains a major cause of cancer-related mortality among women. Breast-conserving surgery (BCS), also known as lumpectomy, is the removal of the cancerous lump and a small margin of surrounding tissue.... Read more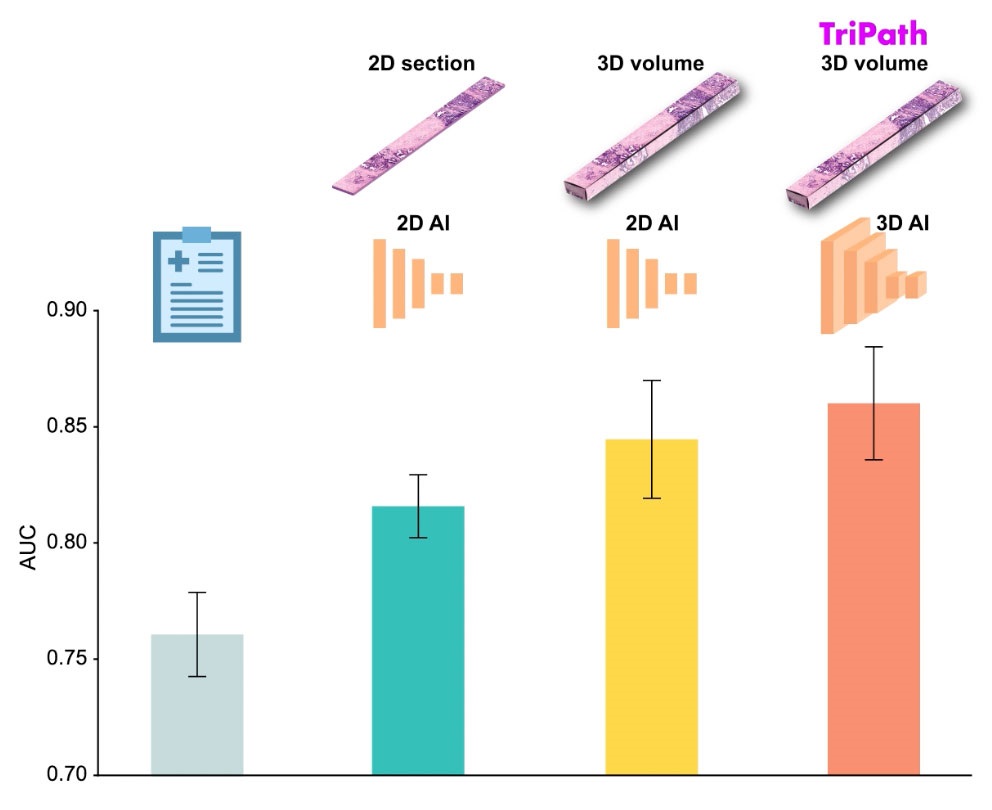
AI Advancements Enable Leap into 3D Pathology
Human tissue is complex, intricate, and naturally three-dimensional. However, the thin two-dimensional tissue slices commonly used by pathologists to diagnose diseases provide only a limited view of the... Read more
New Blood Test Device Modeled on Leeches to Help Diagnose Malaria
Many individuals have a fear of needles, making the experience of having blood drawn from their arm particularly distressing. An alternative method involves taking blood from the fingertip or earlobe,... Read more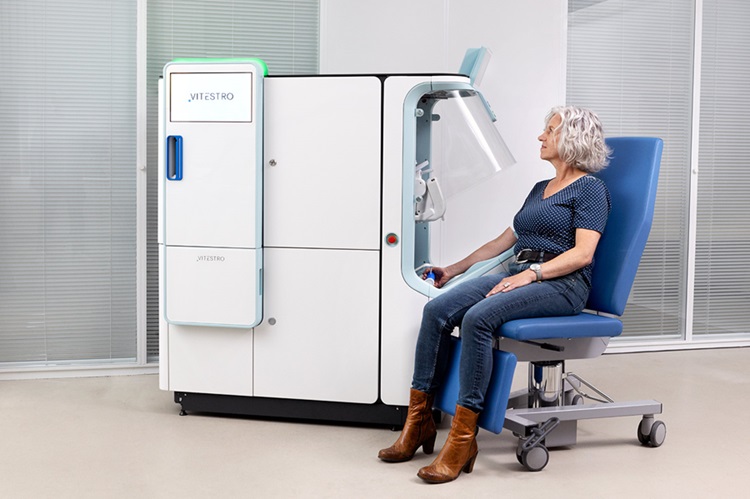
Robotic Blood Drawing Device to Revolutionize Sample Collection for Diagnostic Testing
Blood drawing is performed billions of times each year worldwide, playing a critical role in diagnostic procedures. Despite its importance, clinical laboratories are dealing with significant staff shortages,... Read moreTechnology
view channel
New Diagnostic System Achieves PCR Testing Accuracy
While PCR tests are the gold standard of accuracy for virology testing, they come with limitations such as complexity, the need for skilled lab operators, and longer result times. They also require complex... Read more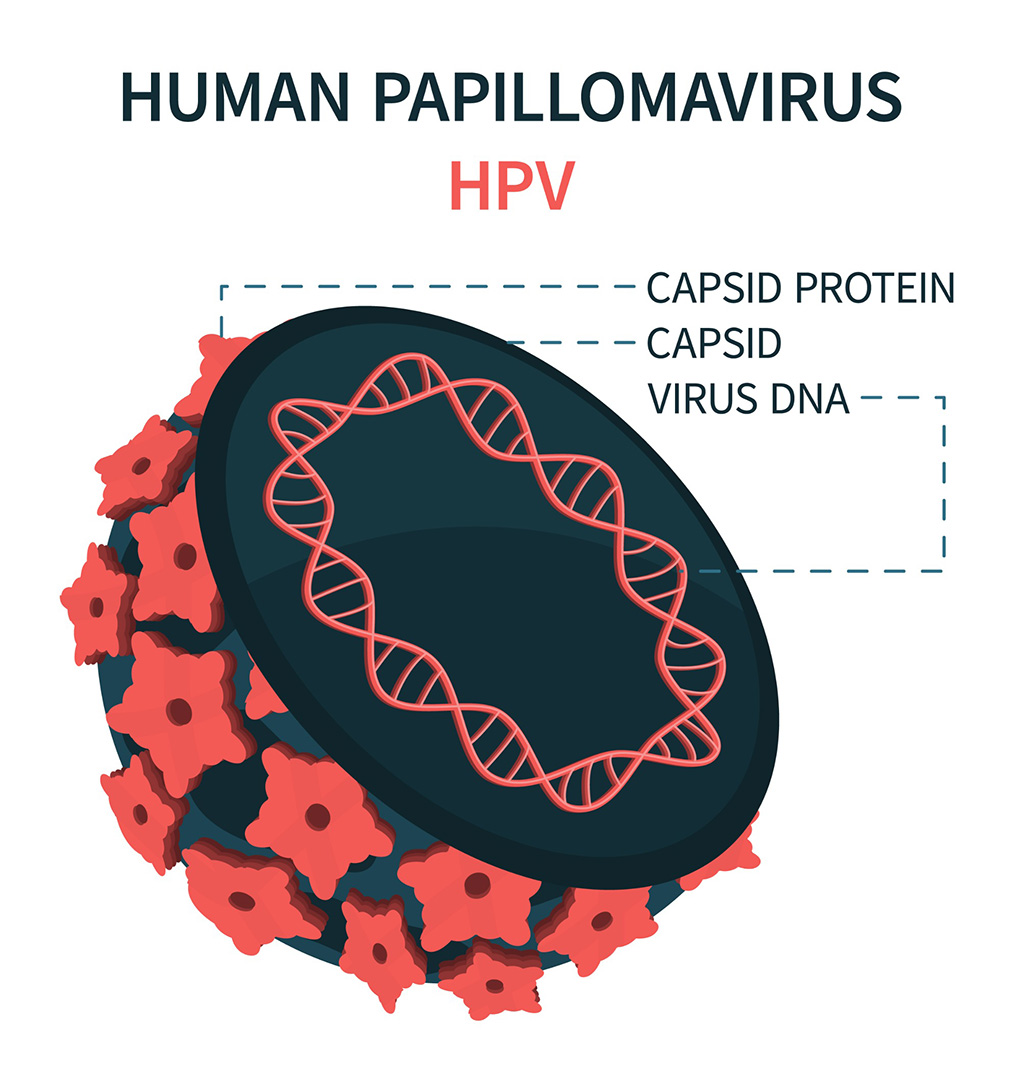
DNA Biosensor Enables Early Diagnosis of Cervical Cancer
Molybdenum disulfide (MoS2), recognized for its potential to form two-dimensional nanosheets like graphene, is a material that's increasingly catching the eye of the scientific community.... Read more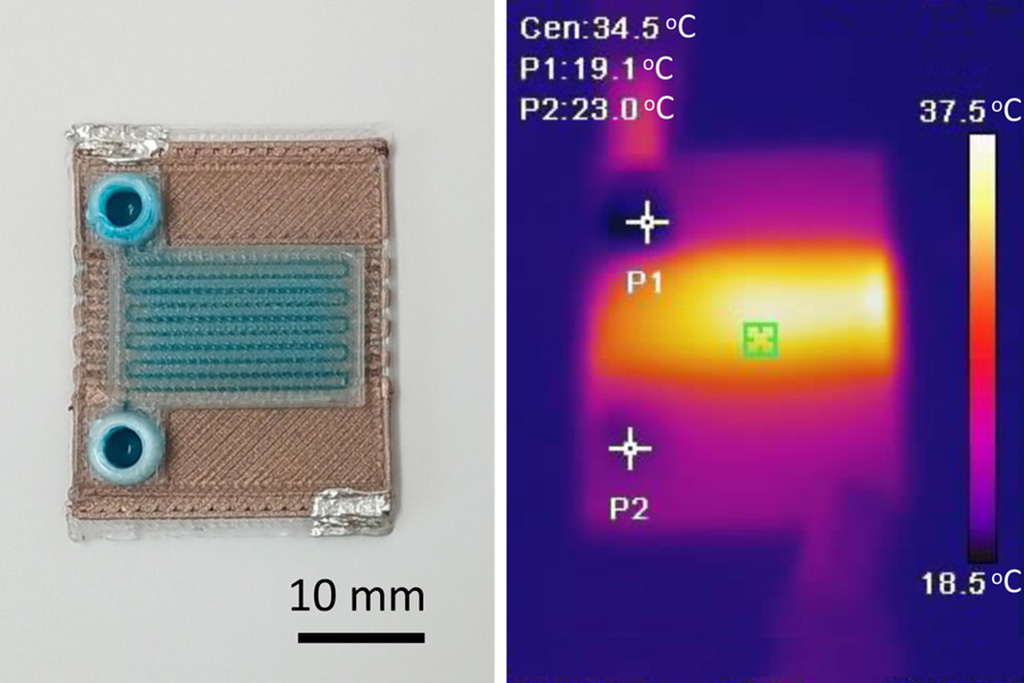
Self-Heating Microfluidic Devices Can Detect Diseases in Tiny Blood or Fluid Samples
Microfluidics, which are miniature devices that control the flow of liquids and facilitate chemical reactions, play a key role in disease detection from small samples of blood or other fluids.... Read more
Breakthrough in Diagnostic Technology Could Make On-The-Spot Testing Widely Accessible
Home testing gained significant importance during the COVID-19 pandemic, yet the availability of rapid tests is limited, and most of them can only drive one liquid across the strip, leading to continued... Read moreIndustry
view channel
Danaher and Johns Hopkins University Collaborate to Improve Neurological Diagnosis
Unlike severe traumatic brain injury (TBI), mild TBI often does not show clear correlations with abnormalities detected through head computed tomography (CT) scans. Consequently, there is a pressing need... Read more
Beckman Coulter and MeMed Expand Host Immune Response Diagnostics Partnership
Beckman Coulter Diagnostics (Brea, CA, USA) and MeMed BV (Haifa, Israel) have expanded their host immune response diagnostics partnership. Beckman Coulter is now an authorized distributor of the MeMed... Read more_1.jpg)










_1.jpg)
.jpg)
