Certain Bacteria Strains Associated with Diabetic Wounds
|
By LabMedica International staff writers Posted on 01 May 2019 |
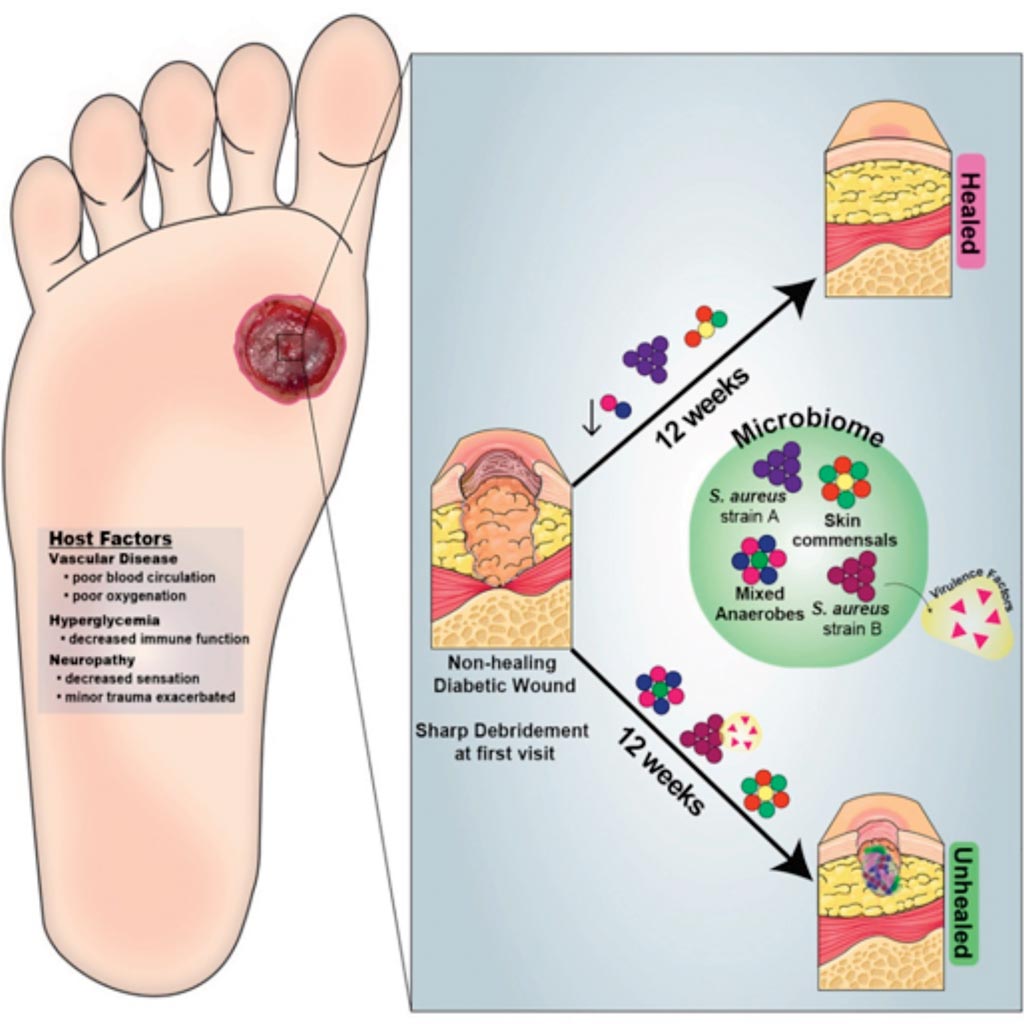
Image: A graphic representation of Strain- and Species-Level Variation in the Microbiome of Diabetic Wounds Is Associated with Clinical Outcomes and Therapeutic Efficacy (Photo courtesy of the University of Pennsylvania).
About 10% of Americans have been diagnosed with diabetes, and one quarter of these patients will develop a wound that does not heal. In the worst case outcome, which occurs in up to 25% percent of these wound-developing patients, the wounds will require an amputation.
Many patients who develop these ulcers may not notice the initial signs, since the high blood glucose of diabetes can lead to a lack of feeling and deformation of the feet. As a result, patients with diabetes commonly develop foot ulcers that may go unnoticed over time. The mortality rate associated with diabetic foot ulcers is equivalent to that of breast cancer and prostate cancer combined, higher than 70% when they lead to amputation.
Scientists at the University of Pennsylvania School of Medicine (Philadelphia, PA, USA) and their colleagues investigated the role of colonizing microbiota in diabetic wound healing, clinical outcomes, and response to interventions, and conducted a longitudinal, prospective study of patients with neuropathic diabetic foot ulcers (DFU). Metagenomic shotgun sequencing revealed that strain-level variation of Staphylococcus aureus and genetic signatures of biofilm formation were associated with poor outcomes.
Cultured wound isolates of S. aureus elicited differential phenotypes in mouse models that corresponded with patient outcomes, while wound “bystanders” such as Corynebacterium striatum and Alcaligenes faecalis, typically considered commensals or contaminants, also significantly impacted wound severity and healing. Antibiotic resistance genes were widespread, and debridement, rather than antibiotic treatment, significantly shifted the DFU microbiota in patients with more favorable outcomes. These findings suggest that the DFU microbiota may be a marker for clinical outcomes and response to therapeutic interventions.
Elizabeth A. Grice, PhD, an associate professor of Dermatology and the lead author of the study, said, “While wounds don't receive the attention of other diseases, they're incredibly common, and our study increases our understanding of how microbes impair or promote healing. It is possible there are bacteria that actually benefit the wound, and we can use what we learned in this study to develop new treatment strategies for non-healing wounds. We hope this study will eventually help identify patients at risk for bad outcomes and lead to treatment innovations that these patients desperately need.” The study was published on April 18, 2019, in the journal Cell Host and Microbe.
Related Links:
University of Pennsylvania School of Medicine
Many patients who develop these ulcers may not notice the initial signs, since the high blood glucose of diabetes can lead to a lack of feeling and deformation of the feet. As a result, patients with diabetes commonly develop foot ulcers that may go unnoticed over time. The mortality rate associated with diabetic foot ulcers is equivalent to that of breast cancer and prostate cancer combined, higher than 70% when they lead to amputation.
Scientists at the University of Pennsylvania School of Medicine (Philadelphia, PA, USA) and their colleagues investigated the role of colonizing microbiota in diabetic wound healing, clinical outcomes, and response to interventions, and conducted a longitudinal, prospective study of patients with neuropathic diabetic foot ulcers (DFU). Metagenomic shotgun sequencing revealed that strain-level variation of Staphylococcus aureus and genetic signatures of biofilm formation were associated with poor outcomes.
Cultured wound isolates of S. aureus elicited differential phenotypes in mouse models that corresponded with patient outcomes, while wound “bystanders” such as Corynebacterium striatum and Alcaligenes faecalis, typically considered commensals or contaminants, also significantly impacted wound severity and healing. Antibiotic resistance genes were widespread, and debridement, rather than antibiotic treatment, significantly shifted the DFU microbiota in patients with more favorable outcomes. These findings suggest that the DFU microbiota may be a marker for clinical outcomes and response to therapeutic interventions.
Elizabeth A. Grice, PhD, an associate professor of Dermatology and the lead author of the study, said, “While wounds don't receive the attention of other diseases, they're incredibly common, and our study increases our understanding of how microbes impair or promote healing. It is possible there are bacteria that actually benefit the wound, and we can use what we learned in this study to develop new treatment strategies for non-healing wounds. We hope this study will eventually help identify patients at risk for bad outcomes and lead to treatment innovations that these patients desperately need.” The study was published on April 18, 2019, in the journal Cell Host and Microbe.
Related Links:
University of Pennsylvania School of Medicine
Latest Microbiology News
- Rapid Sequencing Could Transform Tuberculosis Care
- Blood-Based Viral Signature Identified in Crohn’s Disease
- Hidden Gut Viruses Linked to Colorectal Cancer Risk
- Three-Test Panel Launched for Detection of Liver Fluke Infections
- Rapid Test Promises Faster Answers for Drug-Resistant Infections
- CRISPR-Based Technology Neutralizes Antibiotic-Resistant Bacteria
- Comprehensive Review Identifies Gut Microbiome Signatures Associated With Alzheimer’s Disease
- AI-Powered Platform Enables Rapid Detection of Drug-Resistant C. Auris Pathogens
- New Test Measures How Effectively Antibiotics Kill Bacteria
- New Antimicrobial Stewardship Standards for TB Care to Optimize Diagnostics
- New UTI Diagnosis Method Delivers Antibiotic Resistance Results 24 Hours Earlier
- Breakthroughs in Microbial Analysis to Enhance Disease Prediction
- Blood-Based Diagnostic Method Could Identify Pediatric LRTIs
- Rapid Diagnostic Test Matches Gold Standard for Sepsis Detection
- Rapid POC Tuberculosis Test Provides Results Within 15 Minutes
- Rapid Assay Identifies Bloodstream Infection Pathogens Directly from Patient Samples
Channels
Clinical Chemistry
view channelNew Blood Test Index Offers Earlier Detection of Liver Scarring
Metabolic fatty liver disease is highly prevalent and often silent, yet it can progress to fibrosis, cirrhosis, and liver failure. Current first-line blood test scores frequently return indeterminate results,... Read more
Electronic Nose Smells Early Signs of Ovarian Cancer in Blood
Ovarian cancer is often diagnosed at a late stage because its symptoms are vague and resemble those of more common conditions. Unlike breast cancer, there is currently no reliable screening method, and... Read moreMolecular Diagnostics
view channel
Blood Test Could Spot Common Post-Surgery Condition Early
Heterotopic ossification (HO), the abnormal formation of bone in soft tissue, is a common complication following hip replacement surgery. The condition affects nearly one in three patients and can lead... Read more
New Blood Test Can Help Predict Testicular Cancer Recurrence
Stage 1 testicular germ cell tumor is typically treated with surgery followed by active surveillance. Although most patients experience strong long-term outcomes, about one in four will see their cancer... Read more
New Test Detects Alzheimer’s by Analyzing Altered Protein Shapes in Blood
Alzheimer’s disease begins developing years before memory loss or other symptoms become visible. Misfolded proteins gradually accumulate in the brain, disrupting normal cellular processes.... Read more
New Diagnostic Markers for Multiple Sclerosis Discovered in Cerebrospinal Fluid
Multiple sclerosis (MS) affects nearly three million people worldwide and can cause symptoms such as numbness, visual disturbances, fatigue, and neurological disability. Diagnosing the disease can be challenging... Read moreHematology
view channel
Rapid Cartridge-Based Test Aims to Expand Access to Hemoglobin Disorder Diagnosis
Sickle cell disease and beta thalassemia are hemoglobin disorders that often require referral to specialized laboratories for definitive diagnosis, delaying results for patients and clinicians.... Read more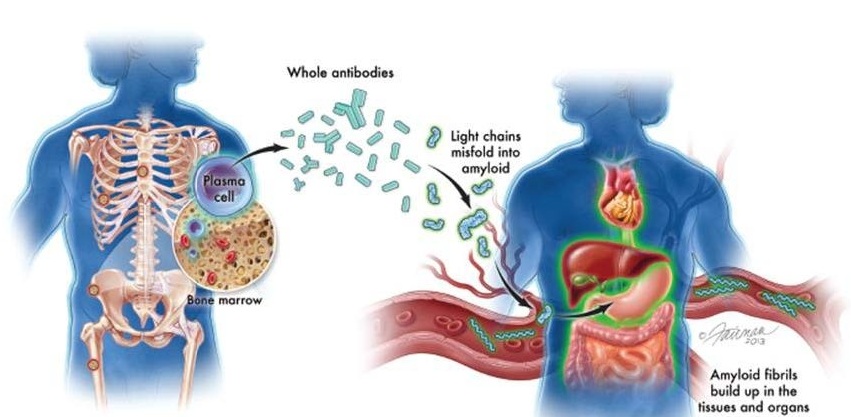
New Guidelines Aim to Improve AL Amyloidosis Diagnosis
Light chain (AL) amyloidosis is a rare, life-threatening bone marrow disorder in which abnormal amyloid proteins accumulate in organs. Approximately 3,260 people in the United States are diagnosed... Read moreImmunology
view channel
Cancer Mutation ‘Fingerprints’ to Improve Prediction of Immunotherapy Response
Cancer cells accumulate thousands of genetic mutations, but not all mutations affect tumors in the same way. Some make cancer cells more visible to the immune system, while others allow tumors to evade... Read more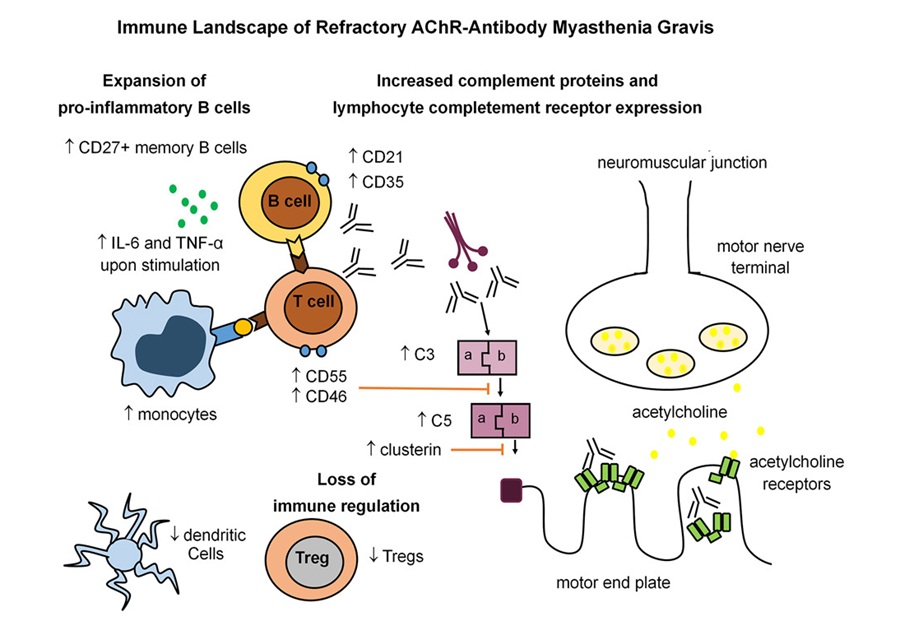
Immune Signature Identified in Treatment-Resistant Myasthenia Gravis
Myasthenia gravis is a rare autoimmune disorder in which immune attack at the neuromuscular junction causes fluctuating weakness that can impair vision, movement, speech, swallowing, and breathing.... Read more
New Biomarker Predicts Chemotherapy Response in Triple-Negative Breast Cancer
Triple-negative breast cancer is an aggressive form of breast cancer in which patients often show widely varying responses to chemotherapy. Predicting who will benefit from treatment remains challenging,... Read moreBlood Test Identifies Lung Cancer Patients Who Can Benefit from Immunotherapy Drug
Small cell lung cancer (SCLC) is an aggressive disease with limited treatment options, and even newly approved immunotherapies do not benefit all patients. While immunotherapy can extend survival for some,... Read morePathology
view channel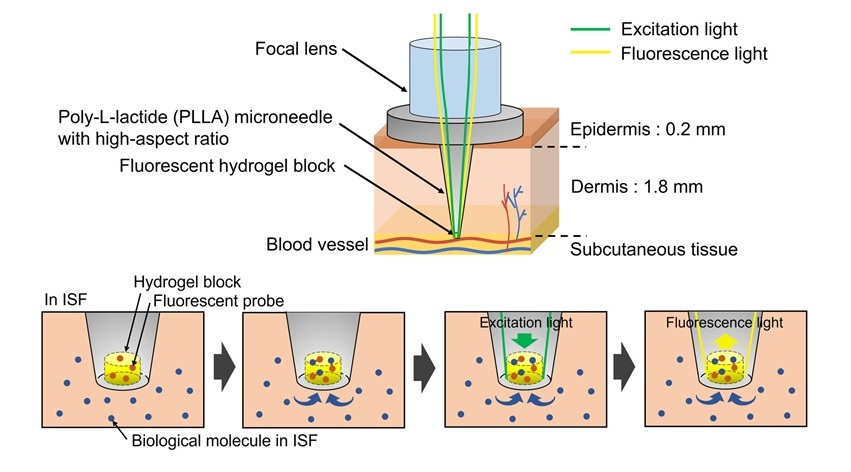
World’s First Optical Microneedle Device to Enable Blood-Sampling-Free Clinical Testing
Blood sampling is one of the most common clinical procedures, but it can be difficult or uncomfortable for many patients, especially older adults or individuals with certain medical conditions.... Read more
Pathogen-Agnostic Testing Reveals Hidden Respiratory Threats in Negative Samples
Polymerase Chain Reaction (PCR) testing became widely recognized during the COVID-19 pandemic as a powerful method for detecting viruses such as SARS-CoV-2. PCR belongs to a group of diagnostic methods... Read moreTechnology
view channel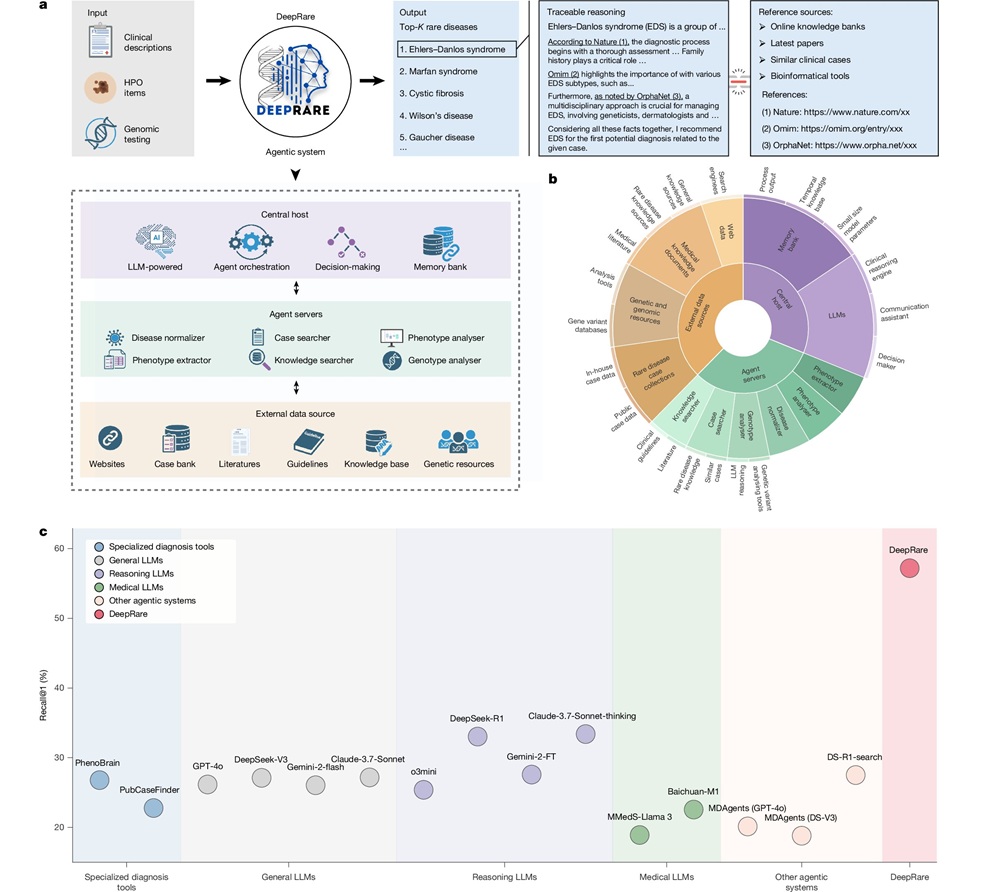
AI Model Outperforms Clinicians in Rare Disease Detection
Rare diseases affect an estimated 300 million people worldwide, yet diagnosis is often protracted and error-prone. Many conditions present with heterogeneous signs that overlap with common disorders, leading... Read more
AI-Driven Diagnostic Demonstrates High Accuracy in Detecting Periprosthetic Joint Infection
Periprosthetic joint infection (PJI) is a rare but serious complication affecting 1% to 2% of primary joint replacement surgeries. The condition occurs when bacteria or fungi infect tissues around an implanted... Read moreIndustry
view channel
Cepheid Joins CDC Initiative to Strengthen U.S. Pandemic Testing Preparednesss
Cepheid (Sunnyvale, CA, USA) has been selected by the U.S. Centers for Disease Control and Prevention (CDC) as one of four national collaborators in a federal initiative to speed rapid diagnostic technologies... Read more
QuidelOrtho Collaborates with Lifotronic to Expand Global Immunoassay Portfolio
QuidelOrtho (San Diego, CA, USA) has entered a long-term strategic supply agreement with Lifotronic Technology (Shenzhen, China) to expand its global immunoassay portfolio and accelerate customer access... Read more