Shotgun Metagenomic Technique Detects Tuberculosis Bacteria in Patient Samples Without Culture or Enrichment
|
By LabMedica International staff writers Posted on 15 Oct 2014 |
Infectious disease researchers have developed a new approach for the diagnosis of tuberculosis (TB) that relies on shotgun metagenomics, a method for direct sequencing of DNA extracted from sputum samples, which detects and characterizes the Mycobacterium that cause TB without the need for time-consuming culture or enrichment.
Metagenomics is the study of genetic material recovered directly from environmental samples. In metagenomic sequencing, DNA is recovered directly from environmental samples in an untargeted manner with the goal of obtaining an unbiased sample from all genes of all members of the community. Recent studies used shotgun Sanger sequencing or pyrosequencing to recover the sequences of the reads. Shotgun sequencing is a sequencing method designed for analysis of DNA sequences longer than 1,000 base pairs, up to and including entire chromosomes. This method requires the target DNA to be broken into random fragments. After sequencing individual fragments, the sequences can be reassembled on the basis of their overlapping regions
Investigators at Warwick Medical School (United Kingdom) explored the potential of shotgun metagenomics to detect and characterize strains from the Mycobacterium tuberculosis complex in smear-positive sputum samples. To this end, they analyzed eight samples obtained from tuberculosis patients from Gambia.
The concentration of DNA present in each extract was determined using the Qubit (Invitrogen Ltd., Paisley, United Kingdom) 2.0 fluorometer and Qubit dsDNA Assay Kits according to the manufacturer’s protocol using the HS (high-sensitivity) or BR (broad-range) kits, depending on the DNA concentration. There was no detectable DNA in the negative control samples with the HS kit, which is sensitive down to 10 picograms per microliter. DNA extracts were diluted to 0.2 nanograms per microliter and were then converted into sequencing libraries using the Illumina (Little Chesterford, United Kingdom) Nextera XT sample preparation kit. The libraries were sequenced on the Illumina MiSeq instrument at the University of Warwick.
Using this methodology, the investigators were able to detect sequences from the M. tuberculosis complex in all eight samples, with coverage of the H37Rv reference genome ranging from 0.002X to 0.7X. By analyzing the distribution of large sequence polymorphisms (deletions and the locations of the insertion element IS6110) and single nucleotide polymorphisms (SNPs), they were able to assign seven of eight metagenome-derived genomes to a species and lineage within the M. tuberculosis complex. Two metagenome-derived mycobacterial genomes were assigned to M. africanum, a species largely confined to West Africa; the others that could be assigned belonged to lineages T, H, or LAM within the clade of "modern" M. tuberculosis strains.
"Laboratory diagnosis of TB using conventional approaches is a long drawn-out process, which takes weeks or months," said senior author Dr. Mark Pallen, professor of microbial genomics at Warwick Medical School. "Plus, relying on laboratory culture means using techniques that date back to the 1880s! Metagenomics using the latest high-throughput sequencing technologies and some smart bioinformatics, allows us to detect and characterize the bacteria that cause TB in a matter of a day or two, without having to grow the bacteria, while also giving us key insights into their genome sequences and the lineages that they belong to. We have provided proof-of-principle here, but we still need to make metagenomics more sensitive and improve our workflows. But, caveats aside, let us celebrate the fact that metagenomics stands ready to document past and present infections, shedding light on the emergence, evolution, and spread of microbial pathogens."
The shotgun metagenomics study was published in the September 23, 2014, online edition of the journal PeerJ.
Related Links:
Warwick Medical School
Invitrogen Ltd.
Illumina
Metagenomics is the study of genetic material recovered directly from environmental samples. In metagenomic sequencing, DNA is recovered directly from environmental samples in an untargeted manner with the goal of obtaining an unbiased sample from all genes of all members of the community. Recent studies used shotgun Sanger sequencing or pyrosequencing to recover the sequences of the reads. Shotgun sequencing is a sequencing method designed for analysis of DNA sequences longer than 1,000 base pairs, up to and including entire chromosomes. This method requires the target DNA to be broken into random fragments. After sequencing individual fragments, the sequences can be reassembled on the basis of their overlapping regions
Investigators at Warwick Medical School (United Kingdom) explored the potential of shotgun metagenomics to detect and characterize strains from the Mycobacterium tuberculosis complex in smear-positive sputum samples. To this end, they analyzed eight samples obtained from tuberculosis patients from Gambia.
The concentration of DNA present in each extract was determined using the Qubit (Invitrogen Ltd., Paisley, United Kingdom) 2.0 fluorometer and Qubit dsDNA Assay Kits according to the manufacturer’s protocol using the HS (high-sensitivity) or BR (broad-range) kits, depending on the DNA concentration. There was no detectable DNA in the negative control samples with the HS kit, which is sensitive down to 10 picograms per microliter. DNA extracts were diluted to 0.2 nanograms per microliter and were then converted into sequencing libraries using the Illumina (Little Chesterford, United Kingdom) Nextera XT sample preparation kit. The libraries were sequenced on the Illumina MiSeq instrument at the University of Warwick.
Using this methodology, the investigators were able to detect sequences from the M. tuberculosis complex in all eight samples, with coverage of the H37Rv reference genome ranging from 0.002X to 0.7X. By analyzing the distribution of large sequence polymorphisms (deletions and the locations of the insertion element IS6110) and single nucleotide polymorphisms (SNPs), they were able to assign seven of eight metagenome-derived genomes to a species and lineage within the M. tuberculosis complex. Two metagenome-derived mycobacterial genomes were assigned to M. africanum, a species largely confined to West Africa; the others that could be assigned belonged to lineages T, H, or LAM within the clade of "modern" M. tuberculosis strains.
"Laboratory diagnosis of TB using conventional approaches is a long drawn-out process, which takes weeks or months," said senior author Dr. Mark Pallen, professor of microbial genomics at Warwick Medical School. "Plus, relying on laboratory culture means using techniques that date back to the 1880s! Metagenomics using the latest high-throughput sequencing technologies and some smart bioinformatics, allows us to detect and characterize the bacteria that cause TB in a matter of a day or two, without having to grow the bacteria, while also giving us key insights into their genome sequences and the lineages that they belong to. We have provided proof-of-principle here, but we still need to make metagenomics more sensitive and improve our workflows. But, caveats aside, let us celebrate the fact that metagenomics stands ready to document past and present infections, shedding light on the emergence, evolution, and spread of microbial pathogens."
The shotgun metagenomics study was published in the September 23, 2014, online edition of the journal PeerJ.
Related Links:
Warwick Medical School
Invitrogen Ltd.
Illumina
Read the full article by registering today, it's FREE! 

Register now for FREE to LabMedica.com and get access to news and events that shape the world of Clinical Laboratory Medicine. 
- Free digital version edition of LabMedica International sent by email on regular basis
- Free print version of LabMedica International magazine (available only outside USA and Canada).
- Free and unlimited access to back issues of LabMedica International in digital format
- Free LabMedica International Newsletter sent every week containing the latest news
- Free breaking news sent via email
- Free access to Events Calendar
- Free access to LinkXpress new product services
- REGISTRATION IS FREE AND EASY!
Sign in: Registered website members
Sign in: Registered magazine subscribers
Latest Microbiology News
- Hidden Gut Viruses Linked to Colorectal Cancer Risk
- Three-Test Panel Launched for Detection of Liver Fluke Infections
- Rapid Test Promises Faster Answers for Drug-Resistant Infections
- CRISPR-Based Technology Neutralizes Antibiotic-Resistant Bacteria
- Comprehensive Review Identifies Gut Microbiome Signatures Associated With Alzheimer’s Disease
- AI-Powered Platform Enables Rapid Detection of Drug-Resistant C. Auris Pathogens
- New Test Measures How Effectively Antibiotics Kill Bacteria
- New Antimicrobial Stewardship Standards for TB Care to Optimize Diagnostics
- New UTI Diagnosis Method Delivers Antibiotic Resistance Results 24 Hours Earlier
- Breakthroughs in Microbial Analysis to Enhance Disease Prediction
- Blood-Based Diagnostic Method Could Identify Pediatric LRTIs
- Rapid Diagnostic Test Matches Gold Standard for Sepsis Detection
- Rapid POC Tuberculosis Test Provides Results Within 15 Minutes
- Rapid Assay Identifies Bloodstream Infection Pathogens Directly from Patient Samples
- Blood-Based Molecular Signatures to Enable Rapid EPTB Diagnosis
- 15-Minute Blood Test Diagnoses Life-Threatening Infections in Children
Channels
Clinical Chemistry
view channel
Electronic Nose Smells Early Signs of Ovarian Cancer in Blood
Ovarian cancer is often diagnosed at a late stage because its symptoms are vague and resemble those of more common conditions. Unlike breast cancer, there is currently no reliable screening method, and... Read more
Simple Blood Test Offers New Path to Alzheimer’s Assessment in Primary Care
Timely evaluation of cognitive symptoms in primary care is often limited by restricted access to specialized diagnostics and invasive confirmatory procedures. Clinicians need accessible tools to determine... Read moreMolecular Diagnostics
view channel
New Blood Test Score Detects Hidden Alcohol-Related Liver Disease
Fatty liver disease affects nearly one in three adults worldwide and can be driven by metabolic conditions such as obesity and diabetes or by excessive alcohol use. In routine care, it is often difficult... Read more
New Blood Test Predicts Who Will Most Likely Live Longer
As people age, it becomes increasingly difficult to determine who is likely to maintain stable health and who may face serious decline. Traditional indicators such as age, cholesterol, and physical activity... Read moreHematology
view channel
Rapid Cartridge-Based Test Aims to Expand Access to Hemoglobin Disorder Diagnosis
Sickle cell disease and beta thalassemia are hemoglobin disorders that often require referral to specialized laboratories for definitive diagnosis, delaying results for patients and clinicians.... Read more
New Guidelines Aim to Improve AL Amyloidosis Diagnosis
Light chain (AL) amyloidosis is a rare, life-threatening bone marrow disorder in which abnormal amyloid proteins accumulate in organs. Approximately 3,260 people in the United States are diagnosed... Read moreImmunology
view channel
New Biomarker Predicts Chemotherapy Response in Triple-Negative Breast Cancer
Triple-negative breast cancer is an aggressive form of breast cancer in which patients often show widely varying responses to chemotherapy. Predicting who will benefit from treatment remains challenging,... Read moreBlood Test Identifies Lung Cancer Patients Who Can Benefit from Immunotherapy Drug
Small cell lung cancer (SCLC) is an aggressive disease with limited treatment options, and even newly approved immunotherapies do not benefit all patients. While immunotherapy can extend survival for some,... Read more
Whole-Genome Sequencing Approach Identifies Cancer Patients Benefitting From PARP-Inhibitor Treatment
Targeted cancer therapies such as PARP inhibitors can be highly effective, but only for patients whose tumors carry specific DNA repair defects. Identifying these patients accurately remains challenging,... Read more
Ultrasensitive Liquid Biopsy Demonstrates Efficacy in Predicting Immunotherapy Response
Immunotherapy has transformed cancer treatment, but only a small proportion of patients experience lasting benefit, with response rates often remaining between 10% and 20%. Clinicians currently lack reliable... Read morePathology
view channel
Urine Specimen Collection System Improves Diagnostic Accuracy and Efficiency
Urine testing is a critical, non-invasive diagnostic tool used to detect conditions such as pregnancy, urinary tract infections, metabolic disorders, cancer, and kidney disease. However, contaminated or... Read more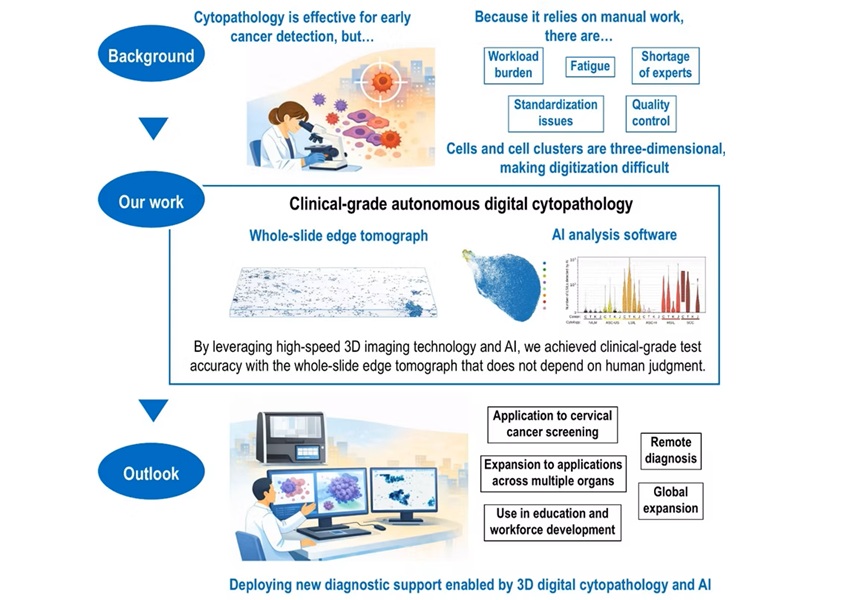
AI-Powered 3D Scanning System Speeds Cancer Screening
Cytology remains a cornerstone of cancer detection, requiring specialists to examine bodily fluids and cells under a microscope. This labor-intensive process involves inspecting up to one million cells... Read moreTechnology
view channel
Blood Test “Clocks” Predict Start of Alzheimer’s Symptoms
More than 7 million Americans live with Alzheimer’s disease, and related health and long-term care costs are projected to reach nearly USD 400 billion in 2025. The disease has no cure, and symptoms often... Read more
AI-Powered Biomarker Predicts Liver Cancer Risk
Liver cancer, or hepatocellular carcinoma, causes more than 800,000 deaths worldwide each year and often goes undetected until late stages. Even after treatment, recurrence rates reach 70% to 80%, contributing... Read more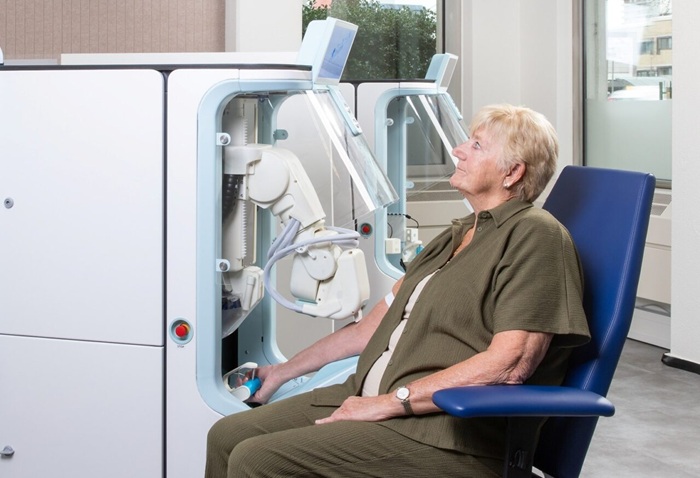
Robotic Technology Unveiled for Automated Diagnostic Blood Draws
Routine diagnostic blood collection is a high‑volume task that can strain staffing and introduce human‑dependent variability, with downstream implications for sample quality and patient experience.... Read more
ADLM Launches First-of-Its-Kind Data Science Program for Laboratory Medicine Professionals
Clinical laboratories generate billions of test results each year, creating a treasure trove of data with the potential to support more personalized testing, improve operational efficiency, and enhance patient care.... Read moreIndustry
view channel
Cepheid Joins CDC Initiative to Strengthen U.S. Pandemic Testing Preparednesss
Cepheid (Sunnyvale, CA, USA) has been selected by the U.S. Centers for Disease Control and Prevention (CDC) as one of four national collaborators in a federal initiative to speed rapid diagnostic technologies... Read more
QuidelOrtho Collaborates with Lifotronic to Expand Global Immunoassay Portfolio
QuidelOrtho (San Diego, CA, USA) has entered a long-term strategic supply agreement with Lifotronic Technology (Shenzhen, China) to expand its global immunoassay portfolio and accelerate customer access... Read more