Lung Adenocarcinoma Genomic Features Differ in East Asians, Europeans
|
By LabMedica International staff writers Posted on 18 Feb 2020 |
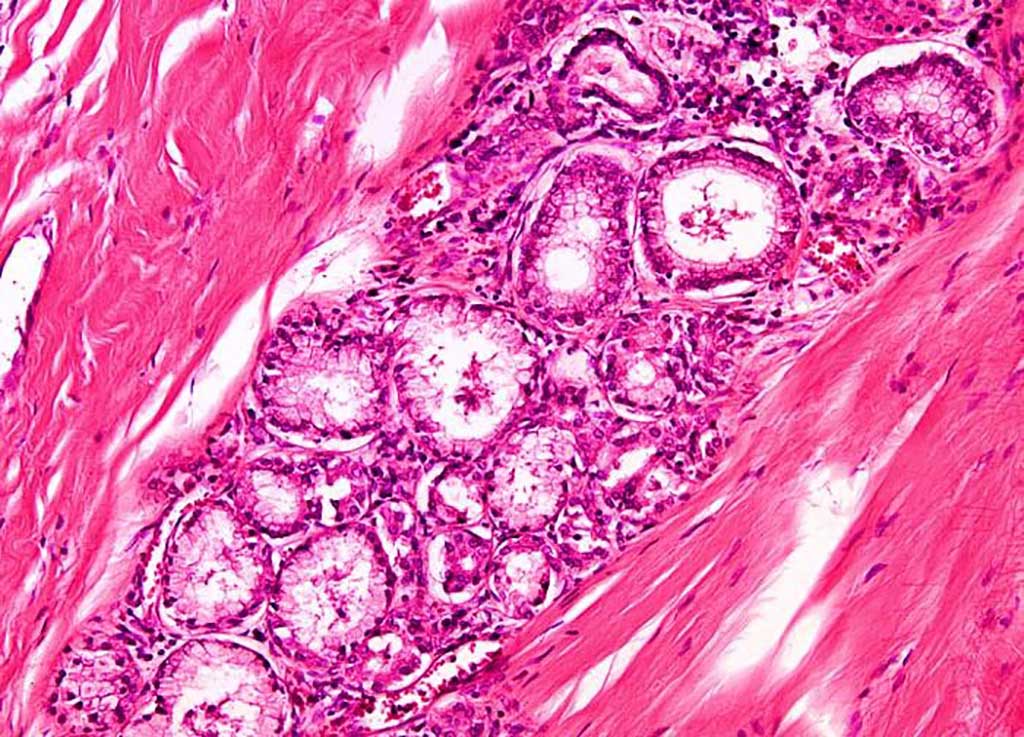
Image: Histopathology of Lung adenocarcinoma (Photo courtesy of Nikon).
Lung adenocarcinoma is a subtype of non-small cell lung cancer (NSCLC). Lung adenocarcinoma is categorized as such by how the cancer cells look under a microscope. Lung adenocarcinoma starts in glandular cells, which secrete substances such as mucus, and tends to develop in smaller airways, such as alveoli.
Lung cancer is the world’s leading cause of cancer death and shows strong ancestry disparities. Lung adenocarcinoma is a common cancer and leads to more than one million deaths each year. A new analysis has found that lung tumors isolated from patients of East Asian ancestry have a less complex genomic architecture than tumors from European patients.
Scientists from the Genome Institute of Singapore (A*STAR, Singapore) and their associates characterized the genomic landscape of lung cancer among East Asians, generating a genomic and transcriptomic dataset encompassing more than 300 lung cancer patients of Chinese ancestry. They sequenced the exomes and transcriptomes of 213 Chinese lung adenocarcinoma patients from Singapore and combined that dataset with previously published whole-exome sequencing data on 92 Chinese patients from another cohort. By comparing the genomic and transcriptomic data from these 305 individuals to that of 249 lung adenocarcinoma patients of European ancestry from The Cancer Genome Atlas, they uncovered differences in tumor mutational burden and driver genes between the groups.
The team reported that overall, East Asian patients' tumors had fewer genomic alterations, with a median tumor mutational burden of 2.04 per Mb, as compared to a median 5.08 per Mb among European patients. While this burden was influenced by patients' smoking status, even among smokers, East Asian patients had a lower median tumor mutational burden than European patients. At the same time, the number and nature of driver mutations differed between tumors from East Asian and European patients. In East Asian patients, alterations affecting the EGFR, TP53, and KRAS genes were the most common driver mutations and nonsmokers had an average 2.08 driver mutations, as compared to an average 2.65 driver mutations among European nonsmokers. Additionally, East Asian patients had fewer copy number variations.
By analyzing the transcriptomic profiles of the tumor samples, the scientists teased out three different lung cancer sub-clusters. Two of these were similar to the terminal respiratory unit (TRU) and proximal inflammatory sub-clusters previously found in European patients, but the third was specific to East Asians. That sub-cluster, dubbed TRU-I, was marked by the upregulation of inflammation-associated genes and increased immune infiltration. This phenotype could help identify patients who might be more likely to benefit from immunotherapy or immune checkpoint blockade treatment.
While they found that patients' clinical features could predict their outcomes, they noted that genomic features could also predict patient survival. These predictions were more accurate for East Asian than European patients, which they attributed to their more stable tumor genomes. The authors concluded that their study elucidated a comprehensive genomic landscape of East Asian ancestry lung adenocarcinomas and highlighted important ancestry differences between the two cohorts. The study was published on February 3, 2020 in the Nature Genetics.
Related Links:
Genome Institute of Singapore
Lung cancer is the world’s leading cause of cancer death and shows strong ancestry disparities. Lung adenocarcinoma is a common cancer and leads to more than one million deaths each year. A new analysis has found that lung tumors isolated from patients of East Asian ancestry have a less complex genomic architecture than tumors from European patients.
Scientists from the Genome Institute of Singapore (A*STAR, Singapore) and their associates characterized the genomic landscape of lung cancer among East Asians, generating a genomic and transcriptomic dataset encompassing more than 300 lung cancer patients of Chinese ancestry. They sequenced the exomes and transcriptomes of 213 Chinese lung adenocarcinoma patients from Singapore and combined that dataset with previously published whole-exome sequencing data on 92 Chinese patients from another cohort. By comparing the genomic and transcriptomic data from these 305 individuals to that of 249 lung adenocarcinoma patients of European ancestry from The Cancer Genome Atlas, they uncovered differences in tumor mutational burden and driver genes between the groups.
The team reported that overall, East Asian patients' tumors had fewer genomic alterations, with a median tumor mutational burden of 2.04 per Mb, as compared to a median 5.08 per Mb among European patients. While this burden was influenced by patients' smoking status, even among smokers, East Asian patients had a lower median tumor mutational burden than European patients. At the same time, the number and nature of driver mutations differed between tumors from East Asian and European patients. In East Asian patients, alterations affecting the EGFR, TP53, and KRAS genes were the most common driver mutations and nonsmokers had an average 2.08 driver mutations, as compared to an average 2.65 driver mutations among European nonsmokers. Additionally, East Asian patients had fewer copy number variations.
By analyzing the transcriptomic profiles of the tumor samples, the scientists teased out three different lung cancer sub-clusters. Two of these were similar to the terminal respiratory unit (TRU) and proximal inflammatory sub-clusters previously found in European patients, but the third was specific to East Asians. That sub-cluster, dubbed TRU-I, was marked by the upregulation of inflammation-associated genes and increased immune infiltration. This phenotype could help identify patients who might be more likely to benefit from immunotherapy or immune checkpoint blockade treatment.
While they found that patients' clinical features could predict their outcomes, they noted that genomic features could also predict patient survival. These predictions were more accurate for East Asian than European patients, which they attributed to their more stable tumor genomes. The authors concluded that their study elucidated a comprehensive genomic landscape of East Asian ancestry lung adenocarcinomas and highlighted important ancestry differences between the two cohorts. The study was published on February 3, 2020 in the Nature Genetics.
Related Links:
Genome Institute of Singapore
Latest Pathology News
- Molecular Imaging to Reduce Need for Melanoma Biopsies
- Urine Specimen Collection System Improves Diagnostic Accuracy and Efficiency
- AI-Powered 3D Scanning System Speeds Cancer Screening
- Single Sample Classifier Predicts Cancer-Associated Fibroblast Subtypes in Patient Samples
- New AI-Driven Platform Standardizes Tuberculosis Smear Microscopy Workflow
- AI Tool Uses Blood Biomarkers to Predict Transplant Complications Before Symptoms Appear
- High-Resolution Cancer Virus Imaging Uncovers Potential Therapeutic Targets
- Research Consortium Harnesses AI and Spatial Biology to Advance Cancer Discovery
- AI Tool Helps See How Cells Work Together Inside Diseased Tissue
- AI-Powered Microscope Diagnoses Malaria in Blood Smears Within Minutes
- Engineered Yeast Cells Enable Rapid Testing of Cancer Immunotherapy
- First-Of-Its-Kind Test Identifies Autism Risk at Birth
- AI Algorithms Improve Genetic Mutation Detection in Cancer Diagnostics
- Skin Biopsy Offers New Diagnostic Method for Neurodegenerative Diseases
- Fast Label-Free Method Identifies Aggressive Cancer Cells
- New X-Ray Method Promises Advances in Histology
Channels
Clinical Chemistry
view channelNew Blood Test Index Offers Earlier Detection of Liver Scarring
Metabolic fatty liver disease is highly prevalent and often silent, yet it can progress to fibrosis, cirrhosis, and liver failure. Current first-line blood test scores frequently return indeterminate results,... Read more
Electronic Nose Smells Early Signs of Ovarian Cancer in Blood
Ovarian cancer is often diagnosed at a late stage because its symptoms are vague and resemble those of more common conditions. Unlike breast cancer, there is currently no reliable screening method, and... Read moreHematology
view channel
Rapid Cartridge-Based Test Aims to Expand Access to Hemoglobin Disorder Diagnosis
Sickle cell disease and beta thalassemia are hemoglobin disorders that often require referral to specialized laboratories for definitive diagnosis, delaying results for patients and clinicians.... Read more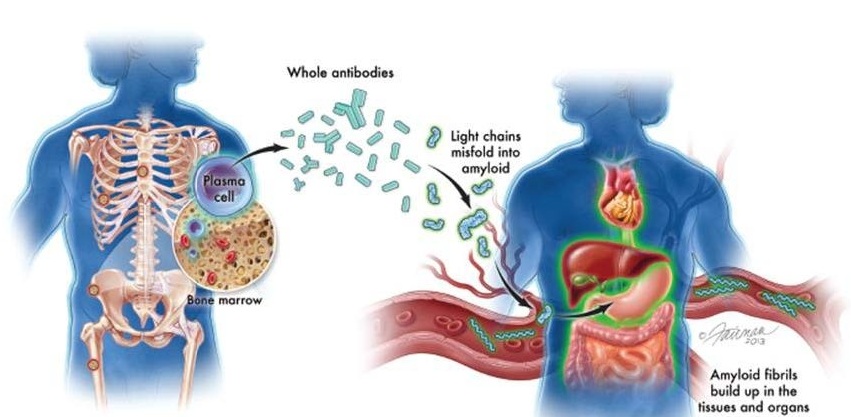
New Guidelines Aim to Improve AL Amyloidosis Diagnosis
Light chain (AL) amyloidosis is a rare, life-threatening bone marrow disorder in which abnormal amyloid proteins accumulate in organs. Approximately 3,260 people in the United States are diagnosed... Read moreImmunology
view channel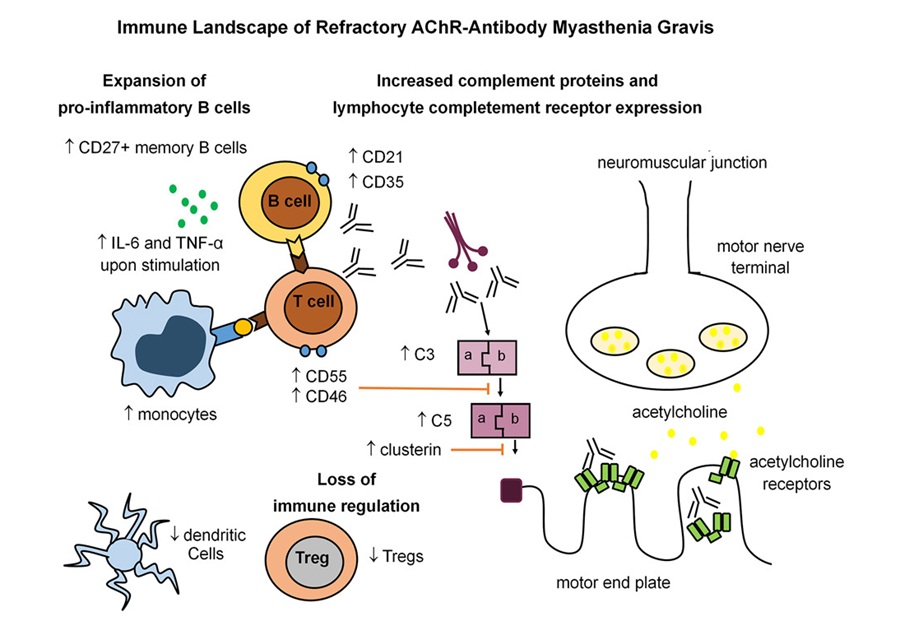
Immune Signature Identified in Treatment-Resistant Myasthenia Gravis
Myasthenia gravis is a rare autoimmune disorder in which immune attack at the neuromuscular junction causes fluctuating weakness that can impair vision, movement, speech, swallowing, and breathing.... Read more
New Biomarker Predicts Chemotherapy Response in Triple-Negative Breast Cancer
Triple-negative breast cancer is an aggressive form of breast cancer in which patients often show widely varying responses to chemotherapy. Predicting who will benefit from treatment remains challenging,... Read moreBlood Test Identifies Lung Cancer Patients Who Can Benefit from Immunotherapy Drug
Small cell lung cancer (SCLC) is an aggressive disease with limited treatment options, and even newly approved immunotherapies do not benefit all patients. While immunotherapy can extend survival for some,... Read more
Whole-Genome Sequencing Approach Identifies Cancer Patients Benefitting From PARP-Inhibitor Treatment
Targeted cancer therapies such as PARP inhibitors can be highly effective, but only for patients whose tumors carry specific DNA repair defects. Identifying these patients accurately remains challenging,... Read moreMicrobiology
view channel
Blood-Based Viral Signature Identified in Crohn’s Disease
Crohn’s disease is a chronic inflammatory intestinal disorder affecting approximately 0.4% of the European population, with symptoms and progression that vary widely. Although viral components of the microbiome... Read more
Hidden Gut Viruses Linked to Colorectal Cancer Risk
Colorectal cancer (CRC) remains a leading cause of cancer mortality in many Western countries, and existing risk-stratification approaches leave substantial room for improvement. Although age, diet, and... Read morePathology
view channel
Molecular Imaging to Reduce Need for Melanoma Biopsies
Melanoma is the deadliest form of skin cancer and accounts for the vast majority of skin cancer-related deaths. Because early melanomas can closely resemble benign moles, clinicians often rely on visual... Read more
Urine Specimen Collection System Improves Diagnostic Accuracy and Efficiency
Urine testing is a critical, non-invasive diagnostic tool used to detect conditions such as pregnancy, urinary tract infections, metabolic disorders, cancer, and kidney disease. However, contaminated or... Read moreTechnology
view channel
AI-Driven Diagnostic Demonstrates High Accuracy in Detecting Periprosthetic Joint Infection
Periprosthetic joint infection (PJI) is a rare but serious complication affecting 1% to 2% of primary joint replacement surgeries. The condition occurs when bacteria or fungi infect tissues around an implanted... Read more
Blood Test “Clocks” Predict Start of Alzheimer’s Symptoms
More than 7 million Americans live with Alzheimer’s disease, and related health and long-term care costs are projected to reach nearly USD 400 billion in 2025. The disease has no cure, and symptoms often... Read moreIndustry
view channel
Cepheid Joins CDC Initiative to Strengthen U.S. Pandemic Testing Preparednesss
Cepheid (Sunnyvale, CA, USA) has been selected by the U.S. Centers for Disease Control and Prevention (CDC) as one of four national collaborators in a federal initiative to speed rapid diagnostic technologies... Read more
QuidelOrtho Collaborates with Lifotronic to Expand Global Immunoassay Portfolio
QuidelOrtho (San Diego, CA, USA) has entered a long-term strategic supply agreement with Lifotronic Technology (Shenzhen, China) to expand its global immunoassay portfolio and accelerate customer access... Read more










 (3) (1).png)