Genotyping Performed by FRET-PCR Without DNA Extraction
|
By LabMedica International staff writers Posted on 07 Jul 2014 |

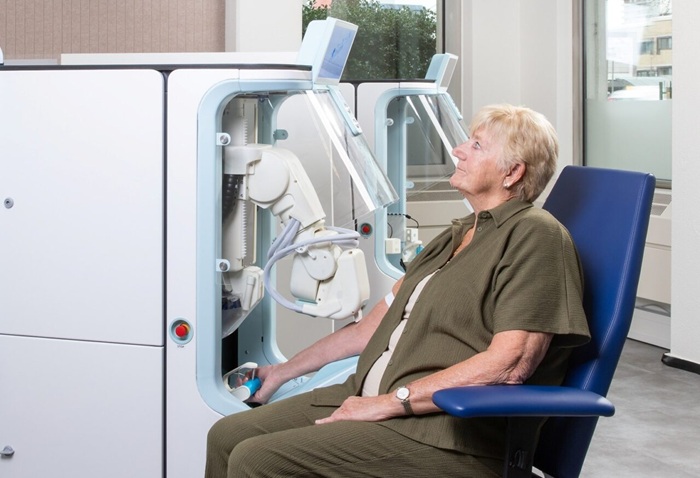
Image: The LightCycler 2.0 real-time polymerase chain reaction analyzer and centrifuge (Photo courtesy of Roche).”
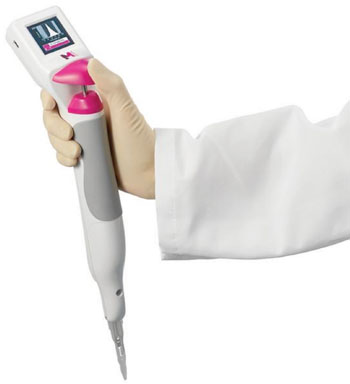
Image: The Scepter 2.0 Automated Cell Counter (Photo courtesy of Merck Millipore).
Blood samples are extensively used for the molecular diagnosis of many hematological diseases using a variety of techniques, based on the amplification of nucleic acids.
Current methods for polymerase chain reaction (PCR) use purified genomic DNA, mostly isolated from total peripheral blood cells or white blood cells (WBC), which can be improved by a real-time fluorescence resonance energy transfer-based method for genotyping directly from blood cells.
Hematologists at the Hospital Universitari Son Espases (Palma de Mallorca, Spain) studied peripheral blood from 34 patients collected into tubes containing ethylenediaminetetraacetic acid (EDTA). Among the samples, they included a mixture of mutant alleles for patients suffering from thrombosis or hereditary hemochromatosis. Red blood cells (RBCs) were lysed and white blood cells (WBCs) isolated. A real-time PCR was then performed followed by a melting curve analysis for different genes including methylenetetrahydrofolate reductase (MTHFR), hemochromatosis (HFE), coagulation factor V Leiden (F5), prothrombin factor two (F2) and coagulation factor XII (F12).
The real time PCR was performed on the LightCycler 2.0 Instrument (Roche Diagnostics Corporation, Indianapolis, IN, USA). In order to standardize the samples for the real-time PCR reaction, cells were counted in a Scepter 2.0 Automated Cell Counter (Merck Millipore, Billerica, MA, USA) and adjusted to 5×106 cells/mL. After testing 34 samples comparing the real-time crossing point (CP) values between 5×106 WBC/mL and 20 ng/µL of purified DNA, the results for F5 Leiden were as follows: CP mean value for WBC was 29.26 ± 0.57 versus purified DNA 24.79 ± 0.56. There was an observed delay of about four cycles when PCR was performed from WBC instead of DNA.
The authors concluded that their protocol obviates the DNA purification stage, thereby saving time and resources. Furthermore, since the manipulation performed on the sample is minimal, it may decrease the risk of contamination. As they reported the results from a variety of genes, they contend that their protocol will be suitable for the genotyping of almost any inherited polymorphism. The study was published on June 25, 2014, in the Journal of Blood Medicine.
Related Links:
Hospital Universitari Son Espases
Roche Diagnostics Corporation
Merck Millipore
Current methods for polymerase chain reaction (PCR) use purified genomic DNA, mostly isolated from total peripheral blood cells or white blood cells (WBC), which can be improved by a real-time fluorescence resonance energy transfer-based method for genotyping directly from blood cells.
Hematologists at the Hospital Universitari Son Espases (Palma de Mallorca, Spain) studied peripheral blood from 34 patients collected into tubes containing ethylenediaminetetraacetic acid (EDTA). Among the samples, they included a mixture of mutant alleles for patients suffering from thrombosis or hereditary hemochromatosis. Red blood cells (RBCs) were lysed and white blood cells (WBCs) isolated. A real-time PCR was then performed followed by a melting curve analysis for different genes including methylenetetrahydrofolate reductase (MTHFR), hemochromatosis (HFE), coagulation factor V Leiden (F5), prothrombin factor two (F2) and coagulation factor XII (F12).
The real time PCR was performed on the LightCycler 2.0 Instrument (Roche Diagnostics Corporation, Indianapolis, IN, USA). In order to standardize the samples for the real-time PCR reaction, cells were counted in a Scepter 2.0 Automated Cell Counter (Merck Millipore, Billerica, MA, USA) and adjusted to 5×106 cells/mL. After testing 34 samples comparing the real-time crossing point (CP) values between 5×106 WBC/mL and 20 ng/µL of purified DNA, the results for F5 Leiden were as follows: CP mean value for WBC was 29.26 ± 0.57 versus purified DNA 24.79 ± 0.56. There was an observed delay of about four cycles when PCR was performed from WBC instead of DNA.
The authors concluded that their protocol obviates the DNA purification stage, thereby saving time and resources. Furthermore, since the manipulation performed on the sample is minimal, it may decrease the risk of contamination. As they reported the results from a variety of genes, they contend that their protocol will be suitable for the genotyping of almost any inherited polymorphism. The study was published on June 25, 2014, in the Journal of Blood Medicine.
Related Links:
Hospital Universitari Son Espases
Roche Diagnostics Corporation
Merck Millipore
Latest Molecular Diagnostics News
- Swab Test Helps Transplant Patients Receive Right Anti-Rejection Medication Dose
- Blood Test Predicts Which Bladder Cancer Patients May Safely Skip Surgery
- Ultra-Sensitive DNA Test Identifies Relapse Risk in Aggressive Leukemia
- Blood Test Could Help Detect Gallbladder Cancer Earlier
- New Blood Test Score Detects Hidden Alcohol-Related Liver Disease
- New Blood Test Predicts Who Will Most Likely Live Longer
- Genetic Test Predicts Radiation Therapy Risk for Prostate Cancer Patients
- Genetic Test Aids Early Detection and Improved Treatment for Cancers
- New Genome Sequencing Technique Measures Epstein-Barr Virus in Blood
- Blood Test Boosts Early Detection of Brain Cancer
- Molecular Monitoring Approach Helps Bladder Cancer Patients Avoid Surgery
- Genetic Tests to Speed Diagnosis of Lymphatic Disorders
- Changes In Lymphatic Vessels Can Aid Early Identification of Aggressive Oral Cancer
- New Extraction Kit Enables Consistent, Scalable cfDNA Isolation from Multiple Biofluids
- New CSF Liquid Biopsy Assay Reveals Genomic Insights for CNS Tumors
- AI-Powered Liquid Biopsy Classifies Pediatric Brain Tumors with High Accuracy
Channels
Clinical Chemistry
view channelNew Blood Test Index Offers Earlier Detection of Liver Scarring
Metabolic fatty liver disease is highly prevalent and often silent, yet it can progress to fibrosis, cirrhosis, and liver failure. Current first-line blood test scores frequently return indeterminate results,... Read more
Electronic Nose Smells Early Signs of Ovarian Cancer in Blood
Ovarian cancer is often diagnosed at a late stage because its symptoms are vague and resemble those of more common conditions. Unlike breast cancer, there is currently no reliable screening method, and... Read moreMolecular Diagnostics
view channel
Swab Test Helps Transplant Patients Receive Right Anti-Rejection Medication Dose
Tacrolimus is widely used to prevent organ rejection in transplant recipients, but achieving the correct dose early is critical. If levels are too low, the transplanted organ may be rejected; if too high,... Read more
Blood Test Predicts Which Bladder Cancer Patients May Safely Skip Surgery
Muscle-invasive bladder cancer (MIBC) often requires removal of the bladder, a procedure that significantly affects quality of life and carries a risk of complications. While newer treatments have improved... Read moreImmunology
view channel
New Biomarker Predicts Chemotherapy Response in Triple-Negative Breast Cancer
Triple-negative breast cancer is an aggressive form of breast cancer in which patients often show widely varying responses to chemotherapy. Predicting who will benefit from treatment remains challenging,... Read moreBlood Test Identifies Lung Cancer Patients Who Can Benefit from Immunotherapy Drug
Small cell lung cancer (SCLC) is an aggressive disease with limited treatment options, and even newly approved immunotherapies do not benefit all patients. While immunotherapy can extend survival for some,... Read more
Whole-Genome Sequencing Approach Identifies Cancer Patients Benefitting From PARP-Inhibitor Treatment
Targeted cancer therapies such as PARP inhibitors can be highly effective, but only for patients whose tumors carry specific DNA repair defects. Identifying these patients accurately remains challenging,... Read more
Ultrasensitive Liquid Biopsy Demonstrates Efficacy in Predicting Immunotherapy Response
Immunotherapy has transformed cancer treatment, but only a small proportion of patients experience lasting benefit, with response rates often remaining between 10% and 20%. Clinicians currently lack reliable... Read moreMicrobiology
view channel
Blood-Based Viral Signature Identified in Crohn’s Disease
Crohn’s disease is a chronic inflammatory intestinal disorder affecting approximately 0.4% of the European population, with symptoms and progression that vary widely. Although viral components of the microbiome... Read more
Hidden Gut Viruses Linked to Colorectal Cancer Risk
Colorectal cancer (CRC) remains a leading cause of cancer mortality in many Western countries, and existing risk-stratification approaches leave substantial room for improvement. Although age, diet, and... Read morePathology
view channel
Molecular Imaging to Reduce Need for Melanoma Biopsies
Melanoma is the deadliest form of skin cancer and accounts for the vast majority of skin cancer-related deaths. Because early melanomas can closely resemble benign moles, clinicians often rely on visual... Read more
Urine Specimen Collection System Improves Diagnostic Accuracy and Efficiency
Urine testing is a critical, non-invasive diagnostic tool used to detect conditions such as pregnancy, urinary tract infections, metabolic disorders, cancer, and kidney disease. However, contaminated or... Read moreTechnology
view channel
AI-Driven Diagnostic Demonstrates High Accuracy in Detecting Periprosthetic Joint Infection
Periprosthetic joint infection (PJI) is a rare but serious complication affecting 1% to 2% of primary joint replacement surgeries. The condition occurs when bacteria or fungi infect tissues around an implanted... Read more
Blood Test “Clocks” Predict Start of Alzheimer’s Symptoms
More than 7 million Americans live with Alzheimer’s disease, and related health and long-term care costs are projected to reach nearly USD 400 billion in 2025. The disease has no cure, and symptoms often... Read moreIndustry
view channel
Cepheid Joins CDC Initiative to Strengthen U.S. Pandemic Testing Preparednesss
Cepheid (Sunnyvale, CA, USA) has been selected by the U.S. Centers for Disease Control and Prevention (CDC) as one of four national collaborators in a federal initiative to speed rapid diagnostic technologies... Read more
QuidelOrtho Collaborates with Lifotronic to Expand Global Immunoassay Portfolio
QuidelOrtho (San Diego, CA, USA) has entered a long-term strategic supply agreement with Lifotronic Technology (Shenzhen, China) to expand its global immunoassay portfolio and accelerate customer access... Read more