New Virus Identified in Blood Supply
|
By LabMedica International staff writers Posted on 30 Sep 2015 |

Image: The HiSeq 2500 sequencing platform (Photo courtesy of Illumina).
Transfusion of blood or blood-derived products can save lives and improve health but requires safety measures for preventing bystander transmission of infectious agents.
To investigate the transmission of novel infectious agents by blood transfusion, changes in the virome composition of blood transfusion recipients pre- and post-transfusion has been studied.
An international group of scientists led by those at Columbia University (New York, NY, USA) collected serum samples from four participating blood centers distributed across the USA from July 1974 through June 1980. They performed high-throughput sequencing on blood samples from 46 individuals in the in the Transfusion-Transmitted Viruses Study (TTVS). They also performed high-throughput sequencing on samples from 106 individuals in the Multicenter Hemophilia Cohort Study who received plasma-derived clotting factor concentrates.
Total ribonucleic acid (RNA) extracts were reverse transcribed using a SuperScript III kit (Invitrogen Life Technologies; Carlsbad, CA, USA) with random hexamer primers. After processing, samples with low concentrations were amplified by increasing polymerase chain reaction (PCR) cycle numbers from 9 to 14. All sequencing was done on the Illumina HiSeq 2500 platform (Illumina; San Diego, CA, USA), yielding an average of 150 million reads per sequencing lane.
The team analyzed samples both pre- and post-transfusion and along with a variety of known viruses, they identified a new virus in two individuals. The virus was only present in post-transfusion samples, and additional tests showed that both patients were able to clear the virus. Genetic analysis determined that the novel human virus, human hepegivirus 1 (HHpgV-1) was related to Hepatitis C virus (HCV) and human Pegivirus (HPgV; formerly called GB virus C or Hepatitis G virus). Genomic testing of 70 additional individuals in the TTVS study failed to detect further cases of the virus detected. HHpgV-1 was found in serum samples from two blood transfusion recipients and two hemophilia patients who had received plasma-derived clotting factor concentrates.
Amit Kapoor, PhD, an assistant professor and lead author of the study said, “HHpgV-1 is unique because it shares genetic similarity with both highly pathogenic HCV and the apparently non-pathogenic HPgV. People need to be aware of this new infection in humans. We just don't know how many viruses are transmitted through the blood supply. There are so many viruses out there, and they need to be characterized in order to ensure that transfusions are safe.” The study was published on September 22, 2015, in the journal mBio.
Related Links:
Columbia University
Invitrogen Life Technologies
Illumina
To investigate the transmission of novel infectious agents by blood transfusion, changes in the virome composition of blood transfusion recipients pre- and post-transfusion has been studied.
An international group of scientists led by those at Columbia University (New York, NY, USA) collected serum samples from four participating blood centers distributed across the USA from July 1974 through June 1980. They performed high-throughput sequencing on blood samples from 46 individuals in the in the Transfusion-Transmitted Viruses Study (TTVS). They also performed high-throughput sequencing on samples from 106 individuals in the Multicenter Hemophilia Cohort Study who received plasma-derived clotting factor concentrates.
Total ribonucleic acid (RNA) extracts were reverse transcribed using a SuperScript III kit (Invitrogen Life Technologies; Carlsbad, CA, USA) with random hexamer primers. After processing, samples with low concentrations were amplified by increasing polymerase chain reaction (PCR) cycle numbers from 9 to 14. All sequencing was done on the Illumina HiSeq 2500 platform (Illumina; San Diego, CA, USA), yielding an average of 150 million reads per sequencing lane.
The team analyzed samples both pre- and post-transfusion and along with a variety of known viruses, they identified a new virus in two individuals. The virus was only present in post-transfusion samples, and additional tests showed that both patients were able to clear the virus. Genetic analysis determined that the novel human virus, human hepegivirus 1 (HHpgV-1) was related to Hepatitis C virus (HCV) and human Pegivirus (HPgV; formerly called GB virus C or Hepatitis G virus). Genomic testing of 70 additional individuals in the TTVS study failed to detect further cases of the virus detected. HHpgV-1 was found in serum samples from two blood transfusion recipients and two hemophilia patients who had received plasma-derived clotting factor concentrates.
Amit Kapoor, PhD, an assistant professor and lead author of the study said, “HHpgV-1 is unique because it shares genetic similarity with both highly pathogenic HCV and the apparently non-pathogenic HPgV. People need to be aware of this new infection in humans. We just don't know how many viruses are transmitted through the blood supply. There are so many viruses out there, and they need to be characterized in order to ensure that transfusions are safe.” The study was published on September 22, 2015, in the journal mBio.
Related Links:
Columbia University
Invitrogen Life Technologies
Illumina
Latest Molecular Diagnostics News
- New CSF Liquid Biopsy Assay Reveals Genomic Insights for CNS Tumors
- Group A Strep Molecular Test Delivers Definitive Results at POC in 15 Minutes
- Rapid Molecular Test Identifies Sepsis Patients Most Likely to Have Positive Blood Cultures
- Light-Based Sensor Detects Early Molecular Signs of Cancer in Blood
- New Testing Method Predicts Trauma Patient Recovery Days in Advance
- Simple Method Predicts Risk of Brain Tumor Recurrence
- Genetic Test Could Improve Early Detection of Prostate Cancer
- Bone Molecular Maps to Transform Early Osteoarthritis Detection
- POC Testing for Hepatitis B DNA as Effective as Traditional Laboratory Testing
- Fully Automated Immunoassay Test Detects HDV Co‑Infection and Super-Infection
- Abdominal Fluid Testing Can Predict Ovarian Cancer Progression
- POC Test Uses Fingerstick Blood, Serum, Or Plasma Sample to Detect Typhoid Fever
- Rapid Testing Panel Simultaneously Detects 15 Drugs of Abuse in Urine Within 21 Minutes
- New Test Detects Breast Reconstruction-Related Infections Before Symptoms Appear
- Period Blood Test for HPV Could Replace Cervical Screening
- New Genetic Tools Improve Breast Cancer Risk Prediction for African American Women
Channels
Clinical Chemistry
view channel
Rapid Blood Testing Method Aids Safer Decision-Making in Drug-Related Emergencies
Acute recreational drug toxicity is a frequent reason for emergency department visits, yet clinicians rarely have access to confirmatory toxicology results in real time. Instead, treatment decisions are... Read more
New PSA-Based Prognostic Model Improves Prostate Cancer Risk Assessment
Prostate cancer is the second-leading cause of cancer death among American men, and about one in eight will be diagnosed in their lifetime. Screening relies on blood levels of prostate-specific antigen... Read moreMolecular Diagnostics
view channel
New CSF Liquid Biopsy Assay Reveals Genomic Insights for CNS Tumors
Central nervous system (CNS) malignancies pose distinctive diagnostic challenges because tissue-based testing is often infeasible and the blood–brain barrier limits the usefulness of plasma liquid biopsy.... Read more
Group A Strep Molecular Test Delivers Definitive Results at POC in 15 Minutes
Strep throat is a bacterial infection caused by Group A Streptococcus (GAS). It is a leading bacterial cause of acute pharyngitis, particularly in children and adolescents, and one of the most common reasons... Read more
Rapid Molecular Test Identifies Sepsis Patients Most Likely to Have Positive Blood Cultures
Sepsis is caused by a patient’s overwhelming immune response to an infection. If undetected or left untreated, sepsis leads to tissue damage, organ failure, permanent disability, and often death.... Read moreHematology
view channel
Rapid Cartridge-Based Test Aims to Expand Access to Hemoglobin Disorder Diagnosis
Sickle cell disease and beta thalassemia are hemoglobin disorders that often require referral to specialized laboratories for definitive diagnosis, delaying results for patients and clinicians.... Read more
New Guidelines Aim to Improve AL Amyloidosis Diagnosis
Light chain (AL) amyloidosis is a rare, life-threatening bone marrow disorder in which abnormal amyloid proteins accumulate in organs. Approximately 3,260 people in the United States are diagnosed... Read moreImmunology
view channelBlood Test Identifies Lung Cancer Patients Who Can Benefit from Immunotherapy Drug
Small cell lung cancer (SCLC) is an aggressive disease with limited treatment options, and even newly approved immunotherapies do not benefit all patients. While immunotherapy can extend survival for some,... Read more
Whole-Genome Sequencing Approach Identifies Cancer Patients Benefitting From PARP-Inhibitor Treatment
Targeted cancer therapies such as PARP inhibitors can be highly effective, but only for patients whose tumors carry specific DNA repair defects. Identifying these patients accurately remains challenging,... Read more
Ultrasensitive Liquid Biopsy Demonstrates Efficacy in Predicting Immunotherapy Response
Immunotherapy has transformed cancer treatment, but only a small proportion of patients experience lasting benefit, with response rates often remaining between 10% and 20%. Clinicians currently lack reliable... Read morePathology
view channel
AI Tool Uses Blood Biomarkers to Predict Transplant Complications Before Symptoms Appear
Stem cell and bone marrow transplants can be lifesaving, but serious complications may arise months after patients leave the hospital. One of the most dangerous is chronic graft-versus-host disease, in... Read more
Research Consortium Harnesses AI and Spatial Biology to Advance Cancer Discovery
AI has the potential to transform cancer care, yet progress remains constrained by fragmented, inaccessible data that hinder advances in early diagnosis and precision therapy. Unlocking patterns missed... Read moreTechnology
view channel
AI-Powered Biomarker Predicts Liver Cancer Risk
Liver cancer, or hepatocellular carcinoma, causes more than 800,000 deaths worldwide each year and often goes undetected until late stages. Even after treatment, recurrence rates reach 70% to 80%, contributing... Read more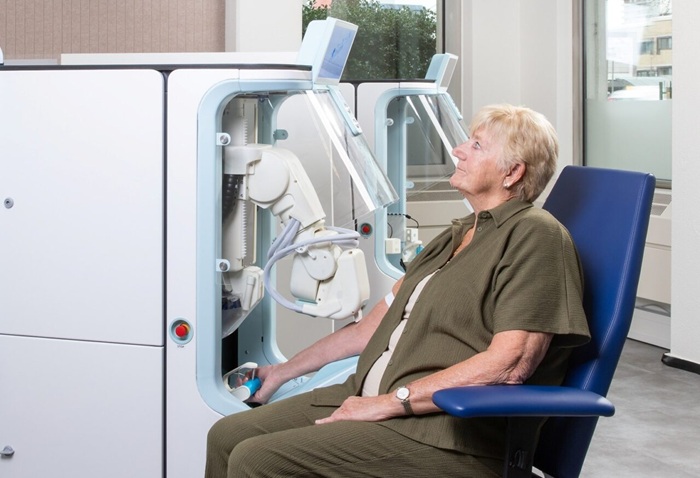
Robotic Technology Unveiled for Automated Diagnostic Blood Draws
Routine diagnostic blood collection is a high‑volume task that can strain staffing and introduce human‑dependent variability, with downstream implications for sample quality and patient experience.... Read more
ADLM Launches First-of-Its-Kind Data Science Program for Laboratory Medicine Professionals
Clinical laboratories generate billions of test results each year, creating a treasure trove of data with the potential to support more personalized testing, improve operational efficiency, and enhance patient care.... Read moreIndustry
view channel
QuidelOrtho Collaborates with Lifotronic to Expand Global Immunoassay Portfolio
QuidelOrtho (San Diego, CA, USA) has entered a long-term strategic supply agreement with Lifotronic Technology (Shenzhen, China) to expand its global immunoassay portfolio and accelerate customer access... Read more