Typhoid Fever Bacteria Sequencing Reveals Drug Resistance Genes
By LabMedica International staff writers
Posted on 21 Mar 2018
Antibiotic resistance is a major problem in Salmonella enterica serovar Typhi, the causative agent of typhoid. Multidrug-resistant (MDR) isolates are prevalent in parts of Asia and Africa and are often associated with the dominant H58 haplotype.Posted on 21 Mar 2018
A typhoid fever outbreak emerged in Pakistan in November 2016, with a number of cases, especially in Sindh Province, resistant to ceftriaxone, a third-generation cephalosporin, and to the first-line drugs chloramphenicol, ampicillin, and trimethoprim-sulfamethoxazole.
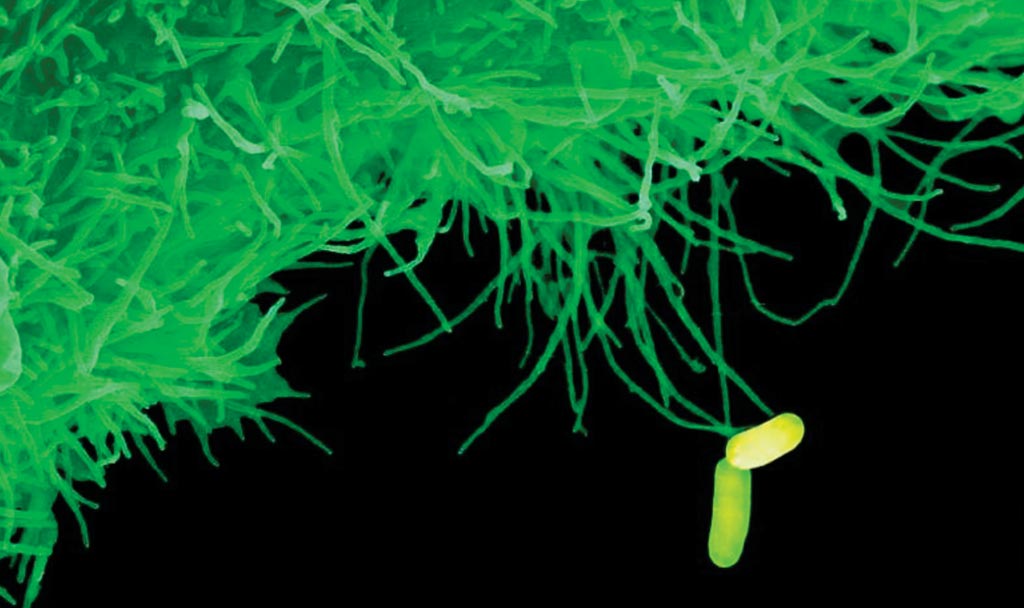
Image: The bacterium Salmonella enterica serovar Typhi in Pakistan have developed greater drug resistance by acquiring a DNA plasmid encoding further antibiotic resistance genes from another bacteria (Photo courtesy of Dave Goulding, Wellcome Sanger Institute).
Scientists collaborating with the Wellcome Trust Sanger Institute (Hinxton, UK) sequenced 80 S. enterica serovar Typhi isolates from this outbreak to find that they harbored resistance genes that are embedded both in chromosomal regions and on a plasmid. The plasmid conferring fluoroquinolone resistance appears to have been acquired from Escherichia coli.
The investigators used the HiSeq 2500 platform to sequence 87 isolates, mostly from Hyderabad and Karachi, that exhibited a pattern of being extensively drug resistant (XDR). For one sample, the team also generated a complete genome using a combination of Oxford Nanopore and Pacific Biosciences long-read sequencing approaches. The final assembled genome was 4.73 million base pairs in length with an 84,492-base-pair long plasmid. At the same time, they analyzed 12 isolates that were susceptible to ceftriaxone. All the XDR and 11 of the 12 ceftriaxone-susceptible samples belong to the S. Typhi H58 clade.
Further phylogenetic analysis with these and additional H58 strains indicated that the XDR samples and four of the ceftriaxone-susceptible samples belonged to their own branch, which was separated other members of the clade by 17 single nucleotide polymorphisms (SNPs), six of which were specific to the XDR samples. By combing through the samples' genomes for antibiotic resistance genes, they found that the XDR isolates contained a transposon, which has been observed previously in multidrug-resistant H58 strains. Genes within the transposon provide resistance to chloramphenicol, ampicillin, trimethoprim-sulfamethoxazole, and streptomycin.
Gordon Dougan, PhD, FMedSci, FRS, a professor and senior author of the study, said, “We have used genetic sequencing to uncover how this particular strain of typhoid became resistant to several key antibiotics. Sporadic cases of typhoid with these levels of antimicrobial resistance have been seen before, but this is the first time we've seen an ongoing outbreak, which is concerning.” The study was published on February 20, 2018, in the journal mBIO.
Related Links:
Wellcome Trust Sanger Institute