Automated Malaria Diagnosis Enhanced by Deep Neural Networks
By LabMedica International staff writers
Posted on 14 Aug 2020
Plasmodium falciparum malaria remains one of the greatest global health burdens with over 228 million cases globally in 2018. In that year there were approximately 405,000 deaths due to malaria worldwide, with the African region accounting for 93% of these deaths, mostly among children. Posted on 14 Aug 2020
Although there are a range of techniques that have been developed for the diagnosis of malaria, conventional light microscopy on Giemsa‐stained thick and thin blood films remains the gold standard. Techniques such as polymerase chain reaction, flow cytometric assay and fluorescence‐dye based approaches lack a universally standardized methodology, present high costs, and require quality control improvement.
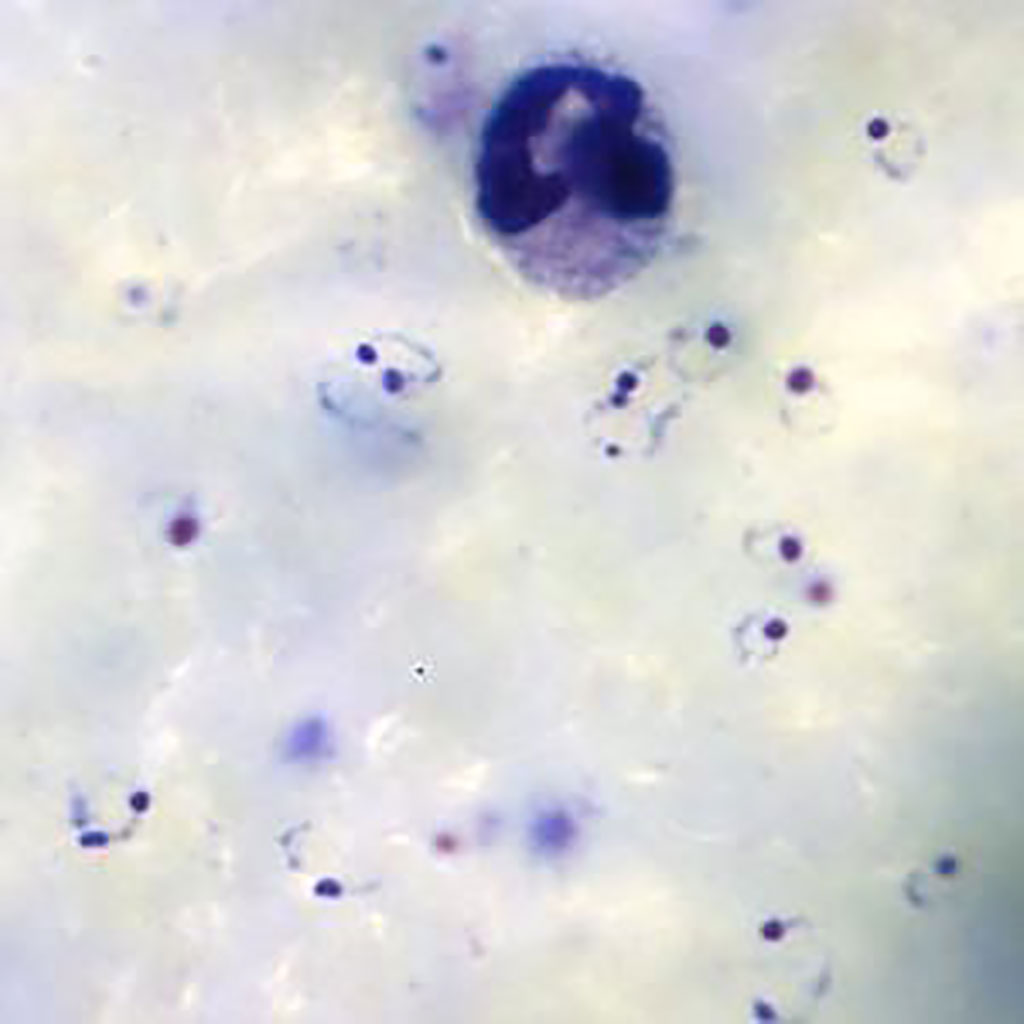
Ring-form trophozoites of Plasmodium falciparum and a white blood cell in a thick blood film (Photo courtesy of Medical Care Development International).
A team of scientists from University College London (London, UK) leveraged routine clinical‐microscopy labels from their quality‐controlled malaria clinics, to train a Deep Malaria Convolutional Neural Network classifier (DeepMCNN) for automated malaria diagnosis. The DeepMCNN system also provides total Malaria Parasite (MP) and White Blood Cell (WBC) counts allowing parasitaemia estimation in MP/μL. Malaria parasites were detected and counted using human‐expert operated microscopy following Giemsa staining of thick and thin blood films. The criterion for declaring a participant to be malaria parasite‐free was no detectable parasites in 100 high‐power (100×) fields in thick films.
The investigators captured images using an upright bright-field BX63 microscope (Olympus, Tokyo, Japan) fitted with a 100×/1.4 NA objective lens, a motorized x‐y sample positioning stage (Prior Scientific, Cambridge, UK) and a color camera to capture images of Giemsa‐stained, thick blood smears. These smears prepared in their clinics tested the use of deep learning‐based object detection methods to identify both P. falciparum parasites and white‐blood‐cell (WBC) nuclei in the digitized extended depth of field (EDoF) thick blood films images.
The team reported that the prospective validation of the DeepMCNN achieved sensitivity/specificity of 0.92/0.90 against expert‐level malaria diagnosis. The PPV/NPV performance was 0.92/0.90, which is clinically usable in their holoendemic settings in a densely populated metropolis.
The authors concluded that their open data and easily deployable DeepMCNN provide a clinically relevant platform, where other healthcare providers could harness their readily available patient level diagnostic labels, to tailor and further improve the accuracy of the DeepMCNN classifier for their clinical pathway settings. The study was published in the August 2020 issue of the American Journal of Hematology.
Related Links:
University College London
Olympus
Prior Scientific










 (3) (1).png)


