Fluorescence-Based Method Detects Misfolded Proteins in Cells
By LabMedica International staff writers
Posted on 25 Feb 2019
A new fluorescence-based method is expected to boost studies on toxic insoluble proteins by detecting aggregation of two proteins in live cells simultaneously.Posted on 25 Feb 2019
Protein aggregation involves the assembly of partially misfolded proteins into oligomeric and higher‐order structures that have been associated with several neurodegenerative diseases. However, numerous questions regarding protein aggregation remain unanswered due to the lack of available tools to visualize these species in living cells.
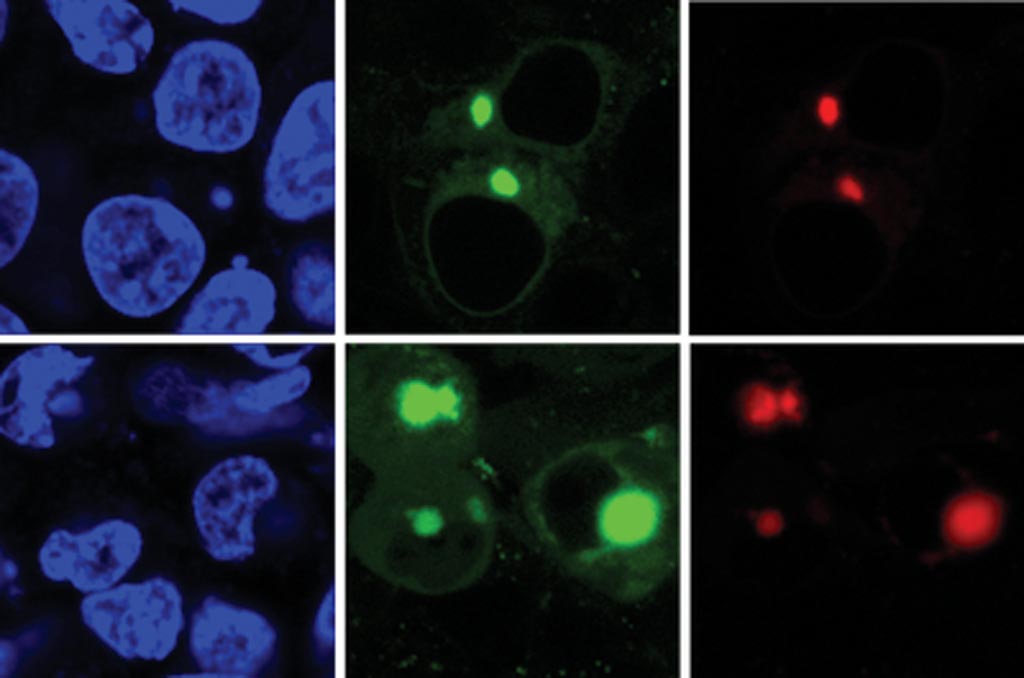
Image: The new AggTag method allows researchers to see the previously undetectable but potentially disease-causing intermediate forms of proteins as they misfold. The method uses fluorescence to simultaneously detect two different proteins (red, green) within the cell (blue) (Photo courtesy of the Zhang Laboratory, Pennsylvania State University).
In this regard, investigators at Pennsylvania State University (University Park, USA) recently (published in the June 8, 2018, online edition of the Journal of the American Chemical Society) developed a fluorogenic method named Aggregation Tag (AggTag), and presented the AggTag probe P1 based on a Halo‐tag ligand to report on the aggregation of a protein of interest in live cells. Halo-tag is a self-labeling protein tag. It is a 297-residue peptide derived from a bacterial enzyme, designed to covalently bind to a synthetic ligand of choice and fuse to a protein of interest. The Halo-tag is composed of two covalently bound segments including a haloalkane dehalogenase and a synthetic ligand of choice. These synthetic ligands consist of a reactive chloroalkane linker bound to a functional group such as biotin or a fluorescent dye, which can be used in the visualization of either living or chemically fixed cells.
Since the Halo‐tag based AggTag method only detected the aggregation of one specific protein of interest at a time, the investigators expanded the AggTag method by using SNAP‐tag technology to enable fluorogenic and biorthogonal detection of the aggregation of two different proteins of interest simultaneously in live cells. A new AggTag probe P2 based on a SNAP‐tag ligand bearing a green fluorescent label was synthesized for this purpose. SNAP-tag is a self-labeling protein tag commercially available in various expression vectors. It is a 182 residues polypeptide that can be fused to any protein of interest and further specifically and covalently tagged with a suitable ligand, such as a fluorescent dye.
The investigators reported in the January 4, 2019, online edition of the journal ChemBioChem that by using confocal imaging and chemical crosslinking experiments, they were able to confirm that P2 could also report on both soluble oligomers and insoluble aggregates of a protein of interest fused with SNAP‐tag in live cells. In addition, they showed that the orthogonal fluorescence of P1 and P2 allowed for simultaneous visualization of two different pathogenic protein aggregates in the same cells.
"In order to function properly, proteins fold into very precise structures, but environmental stress or pathogenic mutations can cause proteins to misfold and aggregate," said senior author Dr. Xin Zhang, assistant professor of chemistry, biochemistry, and molecular biology at Pennsylvania State University. "Protein aggregation is a multi-step process, and it is believed that the intermediate form, which previous imaging techniques could not detect, is responsible for a number of diseases, including Alzheimer's, Parkinson's, Type II diabetes, and cystic fibrosis. We developed the Aggregation Tag method--AggTag--to see these previously undetectable intermediates--soluble oligomers--as well the final aggregates in live cells."
"We plan to continue developing this method so that we can signal the transition of oligomers into insoluble aggregates using a color change," said Dr. Zhang. "This method provides a new toolbox to study protein aggregation, which is currently a highly studied topic among scientists. Hopefully this will allow us to better understand the entire process of protein aggregation and the role of each of these forms in the progression of neurodegenerative and other diseases."
Related Links:
Pennsylvania State University