Protein Identified Modifies Gene Expression in Response to Mitochondrial Damage
By LabMedica International staff writers
Posted on 03 Jan 2017
A protein has been identified that causes changes in cellular gene expression in response to stress or disease related impairment in mitochondrial function.Posted on 03 Jan 2017
Reduced mitochondrial DNA copy number, mitochondrial DNA mutations or disruption of electron transfer chain complexes induce mitochondria-to-nucleus retrograde signaling, which induces global change in nuclear gene expression ultimately contributing to various human pathologies including cancer. Recent studies suggested that these mitochondrial changes caused transcriptional reprogramming of nuclear genes although the mechanism of this cross talk was not clarified.
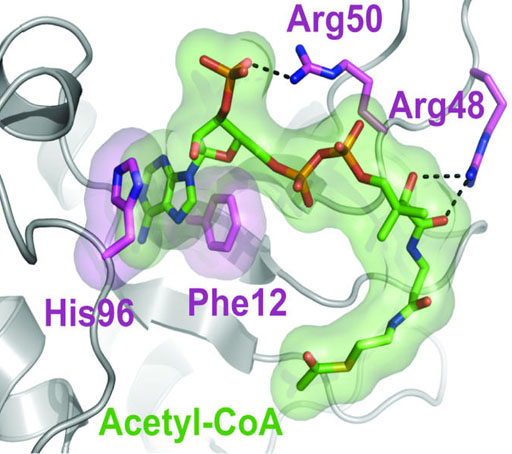
Image: A proposed model of hnRNPA2 (in gray) shows where acetyl-CoA (green) can \"dock\" and interact with different components of the protein. These interactions allow it to transfer acetyl-CoA groups to nuclear gene promoters and translate mitochondrial stress to nuclear gene activation, a process associated with cancer progression (Photo courtesy of the University of Pennsylvania).
Investigators at the University of Pennsylvania (Philadelphia, USA) recently described a mechanism by which mitochondria could drive changes in nuclear gene expression that were associated with tumor progression. They reported in the December 6, 2016, online edition of the journal Cell Discovery that mitochondria-to-nucleus retrograde signaling regulated chromatin acetylation and altered nuclear gene expression through the activity of the protein hnRNAP2 (heterogeneous ribonucleoprotein A2).
To evaluate the role of hnRNPA2 in mitochondria-to-nucleus communication, the investigators traced its activity in cells with depleted levels of mitochondria. Results showed that hnRNPA2 activated gene promoters of stress-associated genes in the nucleus by binding to them. Furthermore, hnRNPA2 activated these genes by acetylating a histone, which loosened the tightly packed chromatin, allowing DNA transcription to proceed more readily. These processes were reversed when mitochondrial DNA content was restored to near normal cell levels.
These findings suggest an important role for mitochondria as determinants of cell fate by altering the expression of large sets of nuclear genes through epigenetic mechanisms.
"Our study provides a rigorous demonstration of a link between mitochondrial function and nuclear gene expression," said senior author Dr. Narayan Avadhani, professor of biochemistry at the University of Pennsylvania. "Since we know that this type of signaling has a direct role in the early stages of cancer progression, the protein involved could be a very valuable target for alleviating this signaling and possibly cancer progression."
Related Links:
University of Pennsylvania








 (3) (1).png)




