Cryoelectron Microscopy Reveals the Ultrastructure of Retroviral Integrase-DNA Complexes
By LabMedica International staff writers
Posted on 29 Feb 2016
Advanced imaging technology has revealed the ultrastructure of the retrovirus intasome, the protein complex that controls the insertion of viral DNA into the host target cell.Posted on 29 Feb 2016
The integrase enzyme catalyses the integration of viral DNA into host target DNA, which is an essential step in the life cycle of all retroviruses. Previous structural characterization of integrase-viral DNA complexes, or intasomes, from the nonpathogenic prototype foamy virus (PFV) revealed a functional integrase tetramer, and it is generally believed that intasomes derived from other retroviral genera use tetrameric integrase. However, the intasomes of orthoretroviruses, which include all known pathogenic species, have not been characterized structurally.
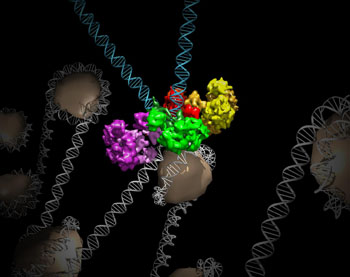
Image: Structure of the intasome protein complex that enables retroviruses to establish permanent infections in their hosts. The intasome hijacks host genomic material, DNA (white) and histones (beige), and irreversibly inserts viral DNA (blue) (Photo courtesy of the Salk Institute for Biological Studies).
Investigators at the Salk Institute for Biological Studies (La Jolla, CA, USA) and Harvard Medical School (Boston, MA, USA) employed the advanced imaging technique cryoelectron microscopy (cryo-EM) to perform structural analysis studies on the pathogenic betaretrovirus, mouse mammary tumor virus (MMTV).
Researchers have historically relied on NMR and X-ray diffraction techniques to determine the structures of molecular complexes and proteins that play a role in the causes of various disease states. Structural information about a variety of medically important proteins and drugs has been obtained by these methods. Cryo-EM is a complementary analytical technique that provides near-atomic resolution without requirements for crystallization or limits on molecular size and complexity imposed by the other techniques. Cryo-EM allows the observation of specimens that have not been stained or fixed in any way, showing them in their native environment while integrating multiple images to form a three-dimensional model of the sample.
Results published in the February 17, 2016, online edition of the journal Nature revealed unexpected octameric integrase architecture for the MMTV. The structure was composed of two core integrase dimers, which interacted with the viral DNA ends and structurally mimicked the PFV integrase tetramer, and two flanking integrase dimers that engaged the core structure via their integrase carboxy-terminal domains. Contrary to the belief that tetrameric integrase components were sufficient to catalyze DNA insertion, the flanking integrase dimers were necessary for MMTV integrase activity.
"The details of how retroviruses integrate differ far more than previously thought and lead to entirely distinct patterns of infection," said contributing author Dr. Dmitry Lyumkis, a research fellow at the Salk Institute for Biological Studies.
"The MMTV intasome structure defines an unexpected novel paradigm for the structural basis of retroviral DNA integration," said senior author Dr. Alan Engelman, professor of medicine at Harvard Medical School.
Related Links:
Salk Institute for Biological Studies
Harvard Medical School