Atomic Structures of Alternative Splicing Regulator Proteins May Lead to New Anticancer Drugs
By LabMedica International staff writers
Posted on 08 Feb 2016
A detailed structural analysis of two RNA-binding proteins that regulate alternate splicing of gene expression, which can lead to various types of cancer, is expected to aid in the development of drugs to suppress this activity.Posted on 08 Feb 2016
Alternative splicing is a regulated process during gene expression that results in a single gene coding for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. Consequently the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions. Notably, alternative splicing allows the human genome to direct the synthesis of many more proteins than would be expected from its 20,000 protein-coding genes.
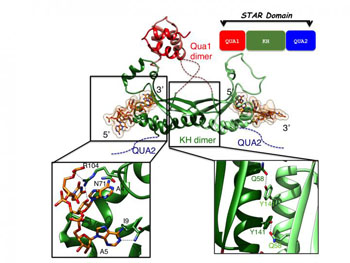
Image: Top: Overview of the structure of T-STAR STAR domain in complex with AUUAAA RNA. Bottom left: close up view of the specific recognition of the RNA. Bottom right: close up view of the KH dimerization interface (Photo courtesy of the University of Leicester).
Investigators at the University of Leicester (United Kingdom) focused on the proteins Sam68 (the Src-Associated substrate in mitosis of 68 kDa) and T-STAR (testis-signal transduction and activation of RNA), which are members of the STAR family of proteins that directly link signal transduction with post-transcriptional gene regulation.
Sam68 is officially called KHDRBS1 (KH domain containing, RNA binding, signal transduction associated 1). It is a KH-type RNA binding protein that recognizes U(U/A)AA direct repeats with relative high affinity (nucleobases A = adenine, U = uracil). Sam68 is predominantly nuclear and its major function in the nucleus is to regulate alternative splicing by recognizing RNA sequences neighboring the included/excluded exon(s). The RNA binding activity of Sam68 is regulated by post-translational modifications such that Sam68 is often referred to as a STAR (Signal Transduction Activator of RNA) protein by which signals from growth factors or soluble tyrosine kinases, such as Src family kinases, act to regulate cellular RNA processes such as alternative splicing. T-STAR is a tissue-specific paralogue (one of a pair of genes that derives from the same ancestral gene and now resides at a different location within the same genome) that regulates the alternative splicing of neuronal pre-mRNAs.
The investigators reported in the January 13, 2016, online edition of the journal Nature Communications that they had determined at atomic resolution how T-STAR and Sam68 bound to RNA, revealing an unexpected mode of dimerization different from other members of the STAR family. They further demonstrated that this unique dimerization interface was crucial for their biological activity in splicing regulation, and suggested that the increased RNA affinity through dimer formation was a crucial parameter enabling these proteins to select their functional targets.
Senior author Dr. Cyril Dominguez, a lecturer in biochemistry at the University of Leicester, said, "The proteins that we have studied, called Sam68 and T-STAR, are very similar, and overexpression of Sam68 has been shown to correlate with poor prognosis in many types of cancers. Our results provide atomic resolution details on how Sam68 binds specifically to its RNA target. Furthermore, we show that Sam68 forms a homodimer that has never been described before and is crucial for its function in RNA splicing. This is important because this basic research set the grounds for structure-based drug design approaches. If we can identify or design drugs that bind specifically at the dimerization interface, we will be able to prevent the function of these proteins in cells, which could have implications for novel cancer treatments. Now that we have a high-resolution structure of Sam68 and T-STAR and a high-throughput binding assay, we are in discussion to collaborate with a major drug discovery consortium to screen a very large library of compounds to inhibit the function of Sam68."
Related Links:
University of Leicester













