One-Tube RNA Ligation and Amplification Method for Rapid Detection of Drug Resistant HIV
By LabMedica International staff writers
Posted on 28 Oct 2015
By not requiring transcription of RNA to DNA, a novel one-tube method allows the rapid detection of drug resistant strains of HIV (human immunodeficiency virus).Posted on 28 Oct 2015
In order to detect point mutations in RNA retroviruses, conventional ligase-mediated approaches require the reverse transcription of viral RNA genomes into DNA before separate ligation and amplification steps can be carried out.
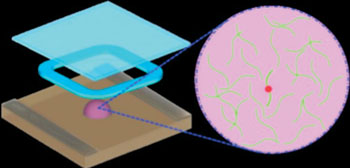
Image: A schematic diagram of a new method that rapidly analyzes the RNA (green strands) of HIV for mutations (red dot) that convey drug resistance. The system does not require transcription of RNA to DNA, as current technologies do, and works within one solution (purple droplet) (Photo courtesy of Dr. Lei Zhang, Brown University).
To simplify this process, investigators at Brown University (Providence, RI, USA) developed one-step ligation on RNA amplification (LRA) method for the direct detection of RNA point mutations. The system operates directly on viral RNA rather than requiring extra, potentially error-prone steps to examine DNA derived from RNA. In a single tube, the system first combines two engineered probes (ligation). If a mutation is present, it then makes many copies of those combined probes (amplification) for detection.
The investigators used this technique for the detection of a common, clinically relevant HIV-1 reverse transcriptase drug-resistant point mutation, K103N, and compared it with allele-specific PCR and pyrosequencing methodology.
They reported in the November 2015 issue of the Journal of Molecular Diagnostics that the LRA test was sensitive enough to detect the K103N mutation in concentrations as low as one mutant per 10,000 strands of normal viral RNA. The LRA test required about two hours while the alternative technologies took as long as eight hours.
"LRA (ligation on RNA amplification) uniquely optimizes two enzymatic reactions—RNA-based ligation, and quantitative PCR (polymerase chain reaction) amplification—into a single system," said senior author Dr. Anubhav Tripathi, professor of engineering at Brown University. "Each HIV contains about 10,000 nucleotides, or building blocks, in its genetic material, and a drop of blood from a patient with resistant HIV can contain thousands to millions of copies of HIV. To find that one virus, out of thousands to millions, which is mutated at just a single nucleotide is like finding a needle in a haystack."
So far the LRA test has been shown to work on RNA that was derived from laboratory HIV strains, but it has not yet been applied to samples from circulating viruses from AIDS patients.
Related Links:
Brown University