Switch to Glycolysis Puts Cells on Road to Becoming Cancerous
By LabMedica International staff writers
Posted on 20 Jul 2015
Mutations that impair the function of the electron transport chain component cytochrome c oxidase (CcO) induce cancerous changes in normal cells and make cancer cells more invasive.Posted on 20 Jul 2015
The enzyme cytochrome c oxidase is a large transmembrane protein complex found in bacteria and the mitochondrion of eukaryotes. It is the last enzyme in the respiratory electron transport chain of mitochondria or bacteria located in the mitochondrial or bacterial membrane. It receives an electron from each of four cytochrome c molecules, and transfers them to one oxygen molecule, converting molecular oxygen to two molecules of water. In the process, it binds four protons from the inner aqueous phase to make water, and in addition translocates four protons across the membrane, helping to establish a transmembrane difference of proton electrochemical potential that ATP synthase then uses to synthesize ATP.
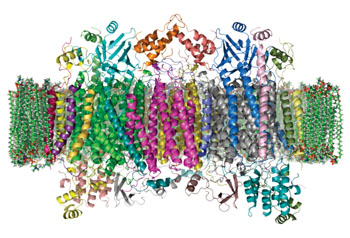
Image: The crystal structure of cytochrome c oxidase in a phospholipid bilayer. The intermembrane space lies to top of the image(Photo courtesy of Wikimedia Commons).
Investigators at the University of Pennsylvania (Philadelphia, USA) used shRNA (short hairpin RNA) to induce genetic silencing of the CcO complex and loss of CcO activity in multiple cell types from mouse and human sources.
They reported in the July 6, 2015, online edition of the journal Oncogene that loss of CcO activity resulted in metabolic shift to glycolysis, loss of anchorage-dependent growth, and acquired invasive phenotypes. Whole-genome expression analysis showed the upregulation of genes involved in cell signaling, extracellular matrix interactions, cell morphogenesis, cell motility, and migration.
The possible use of CcO dysfunction as a biomarker for cancer progression was supported by data indicating that esophageal tumors from human patients showed reduced CcO subunits IVi1 and Vb in regions that were previously found to be located at the hypoxic core of the tumors.
"Disrupting only a single protein subunit of cytochrome oxidase C led to major changes in the mitochondria and in the cells themselves," said senior author Dr. Narayan G. Avadhani, professor of biochemistry at the University of Pennsylvania.
"These cells showed all the characteristics of cancer cells. They displayed changes in their metabolism, becoming more reliant on glucose and reducing their synthesis of ATP. Instead of conducting oxidative phosphorylation, they largely switched over to conducting glycolysis, a less efficient means of making ATP that is common in cancer cells."
"That result alone could not tell us whether that was the cause or effect of tumors, but our cell system clearly says that mitochondrial dysfunction is a driving force in tumorigenesis," said Dr. Avadhani. "We are targeting the signaling pathway, developing a lot of small molecules and antibodies. Hopefully, if you block the signaling, the cells will not go into the so called oncogenic mode and instead would simply die."
Related Links:
University of Pennsylvania