Seeding Assay May Boost Search for Huntington's Disease Drugs
By LabMedica International staff writers
Posted on 08 Jul 2015
An assay technique that measures seeding of mutant huntingtin aggregates in cerebral spinal fluid (CSF) may identify individuals with Huntington's disease (HD) that have not yet become symptomatic and may be used to evaluate the efficacy of new drug treatments for the disease.Posted on 08 Jul 2015
Huntington’s disease is caused by a dominant gene that encodes a protein known as huntingtin (Htt). The 5' end of the Huntington's disease gene has a sequence of three DNA bases, cytosine-adenine-guanine (CAG), coding for the amino acid glutamine, that is repeated multiple times. Normal persons have a CAG repeat count of between seven and 35 repeats, while the mutated form of the gene has anywhere from 36 to 180 repeats. The mutant form of Htt is broken down into toxic peptides, which contribute to the pathology of the syndrome.
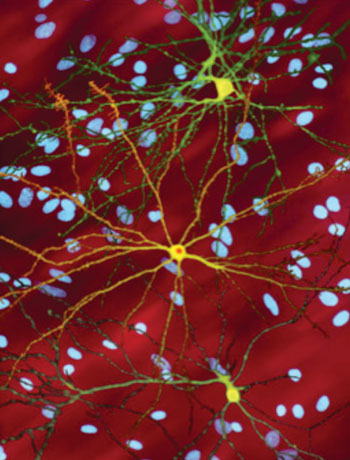
Image: A photmicrograph of medium spiny neurons (yellow) with nuclear inclusions (orange), which occur as part of the Huntington\'s disease process. Several neurons are colored yellow and have a large central core with up to two dozen tendrils branching out of them. The core of the neuron in the foreground contains an orange blob about a quarter of its diameter (Photo courtesy of Wikimedia Commons).
Investigators at the University of California, Irvine (USA) reported in the June 23, 2015, online edition of the journal Molecular Psychiatry that synthetic polyglutamine oligomers and CSF from BACHD transgenic rats and from human HD subjects can seed mutant huntingtin aggregation in a cell model and its cell lysate.
Studies described in the paper demonstrated that seeding required the mutant huntingtin template and may have reflected an underlying prion-like protein propagation mechanism. Light and cryo-electron microscopy showed that synthetic seeds nucleated and enhanced mutant huntingtin aggregation. Seeding was dose dependent, and extracellular seeds were incorporated into endogenously expressed mutant Htt species, forming aggregated fibrils.
The seeding assay distinguished HD subjects from healthy and non-HD dementia controls without overlap. Ultimately, this seeding property in the CSF of HD patient may form the basis of a molecular biomarker assay to monitor HD and evaluate therapies that target mutant Htt.
"Determining if a treatment modifies the course of a neurodegenerative disease like Huntington's or Alzheimer's may take years of clinical observation," said senior author Dr. Steven Potkin, professor of psychiatry and human behavior at the University of California, Irvine. "This assay that reflects a pathological process can play a key role in more rapidly developing an effective treatment. Blocking the cell-to-cell seeding process itself may turn out to be an effective treatment strategy."
Related Links:
University of California, Irvine