Real-Time Studies Show How an Effector Protein Regulates Cellular Gene Expression
By LabMedica International staff writers
Posted on 30 Jun 2015
A series of elegant single molecule experiments have revealed in real-time how the effector protein HP1alpha interacts with chromatin in its target histones to regulate gene expression.Posted on 30 Jun 2015
Cells regulate gene activity at any given time by unwinding specific areas of the packed chromatin (histone-DNA) complex. Conversely, the cell can silence genes by forming compact and dense chromatin fibers that prevent gene expression. This remodeling is carried out by "effector" proteins, which attach to chromatin and change the three-dimensional structure of the gene of interest to either an open or compact format.
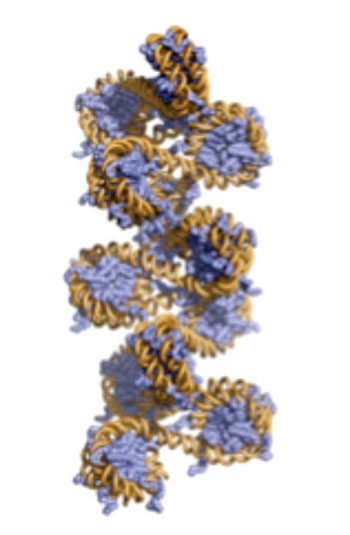
Image: Three-dimensional model of chromatin (Photo courtesy of Dr. Beat Fierze, Ecole Polytechnique Fédérale de Lausanne).
The effector protein HP1alpha is an isoform of the heterochromatin protein 1 (HP1), which plays a role in chromatin remodeling and transcriptional silencing. The HP1 chromo domain selectively recognizes the methylated histones H3-K9, where this methylation is carried out by histone methyltransferase SUV39H1.
Investigators at the Ecole Polytechnique Fédérale de Lausanne (Switzerland) established a fully chemically defined assay system to directly observe the real-time interaction dynamics of individual HP1alpha molecules with their target posttranslational modification (PTMs) histone sites within their native context, nucleosome arrays. This approach was highly complementary to measurements of chromatin effector dynamics in cells, as the investigators maintained complete control over the chromatin architecture and modification status.
Employing chemically defined nucleosome arrays together with single-molecule total internal reflection fluorescence microscopy (smTIRFM), the investigators demonstrated in a paper published in the June 18, 2015, online edition of the journal Nature Communications that HP1alpha residence time on chromatin depended on the density of H3K9me3 histone, as dissociated factors could rapidly rebind at neighboring sites. Moreover, by chemically controlling HP1alpha dimerization they found that effector multivalency prolonged chromatin retention and, importantly, accelerated the association rate.
Senior author Dr. Beat Fierz, professor of biophysical chemistry of macromolecules at Ecole Polytechnique Fédérale de Lausanne, said, "We want to observe complex biology as it happens, and understand it on a quantitative level. Our chemical methods give us complete control over protein-chromatin dynamics, and the current study sets the stage for such unprecedented insights."
Related Links:
Ecole Polytechnique Fédérale de Lausanne