Scientists Unwind More Mysteries of the Cellular Clock
By LabMedica International staff writers
Posted on 09 Dec 2014
A new study surveying circadian gene expression provides a new understanding of how one clock leads to at least 8 different subclocks, and may suggest a new venue for drug treatment of clock-based metabolic disorders.Posted on 09 Dec 2014
Human existence is basically circadian in that most people wake in the morning, sleep in the evening, and eat in between. Body temperature, metabolism, and hormone levels all fluctuate throughout the day and disruption of those cycles can lead to disorders and diseases. The wheels and springs of the molecular clock underlying these circadian rhythms include transcription factors (TFs) that control oscillation of expression of circadian genes.
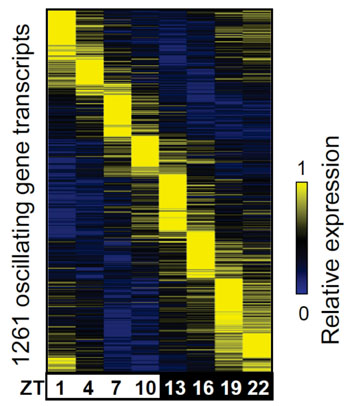
Image: Color coding of genes activated in the liver (yellow) as waves of their activation relative to the time of day. Zeitbeger times (ZT): ZT0= 7 am, ZT3=10 am, ZT6=1 pm, etc. (Photo courtesy of Dr. Bin Fang, Perelman School of Medicine, University of Pennsylvania).
Not all circadian cycles peak at the same time – some peak in the morning, others in the evening, etc. The question is how does a single clock keep time in multiple phases at once?
New findings come from the team of Mitchell Lazar, MD, PhD, and professor at the University of Pennsylvania’s Perelman School of Medicine (Philadelphia, PA, USA) in a report of a genome-wide survey of circadian protein-coding genes and gene-expression enhancers, parts of the “dark matter” of the genome in that they don’t encode for proteins. The team took advantage of new tools based on high-density DNA sequencing to measure the activity of enhancers throughout the day in the livers of mice. They found that many enhancers, like circadian genes themselves, have a daily oscillation that is in phase with nearby genes – both the enhancer and gene activity peak at the same time each day. The activities of the enhancers themselves are, in turn, governed by distinct TFs.
Grouping the enhancers into 8, 3-hour phases based on when they peak, the team asked which TFs are capable of binding to the enhancers in each set. Remarkably, the team found that enhancers that are in the same phase tend to be bound by the same TFs. For instance, one circadian component, CLOCK, binds enhancers that are most active during one particular 3-hour period. Another protein, Rev-erba, binds 12 hours later. Loss of any one of the TFs dysregulates a subset of oscillating genes while leaving genes tuned to other circadian phases unaffected.
“Different transcription factors control different phases,” said Lazar, “That explains how one clock leads to at least 8 different subclocks.”
Also important is that genes that oscillate with the same phase tend to have related functions, which are distinct from the pathways controlled at other times. Genes involved in insulin signaling peak at a different time than genes that control sugar metabolism, for instance.
Lazar suggests it might be possible to tweak pharmaceutical regimens to make them more efficient by delivering drugs only when the pathways they impact are actually active – a strategy that could minimize unintended side effects. Toward “a proof-of-concept for a new principle of drug treatment for metabolic disorders,” he said, “this is a real step in that direction.”
The study, by Fang B, Everett L, Jager J, et al., was published in the journal Cell, online ahead of print November 20, 2014.
Related Links:
Perelman School of Medicine at the University of Pennsylvania
University of Pennsylvania Health System