Advanced Electron Microscopy Reveals Fine Structure of Active Signaling Complexes
By LabMedica International staff writers
Posted on 07 Jul 2014
A team of molecular biologists used advanced electron microscopy and mass spectroscopy techniques to determine the structure of the functional human beta2AR (beta2 adrenergic receptor)-beta-arrestin-1 signaling complex.Posted on 07 Jul 2014
Members of arrestin/beta-arrestin protein family are thought to participate in agonist-mediated desensitization of G-protein-coupled receptors (GPCRs) and cause specific dampening of cellular responses to stimuli such as hormones, neurotransmitters, or sensory signals.
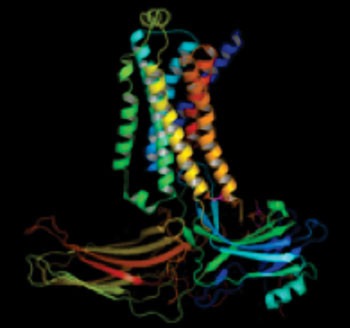
Image: A structural model of the beta2 adrenergic receptor-arrestin signaling complex as deduced by electron microscopy, cross-linking, and mass spectrometry (Photo courtesy of Duke University).
While a recent barrage of structural data on a number of GPCRs including the beta2AR–G-protein complex has provided novel insights into the structural basis of receptor activation, information has been lacking on the recruitment of beta-arrestins to activated GPCRs, primarily owing to challenges in obtaining stable receptor–beta-arrestin complexes for structural studies.
Investigators at Duke University (Durham, NC, USA) and colleagues at the University of Michigan (Ann Arbor, USA) and Stanford University (Palo Alto, CA, USA) devised a strategy for forming and purifying a functional human beta2AR–beta-arrestin-1 complex that allowed them to visualize its architecture by single-particle negative-stain electron microscopy and to characterize the interactions between beta2AR and beta-arrestin 1 using hydrogen–deuterium exchange mass spectrometry (HDX-MS) and chemical cross linking.
Electron microscopy two-dimensional averages and three-dimensional reconstructions revealed bimodal binding of beta-arrestin 1 to the beta2AR, involving two separate sets of interactions, one with the phosphorylated carboxyl terminus of the receptor and the other with its seven-transmembrane core. Areas of reduced HDX together with identification of cross linked residues suggested engagement of the finger loop of beta-arrestin 1 with the seven-transmembrane core of the receptor.
A molecular model of the beta2AR–beta-arrestin signaling complex was made by docking activated beta-arrestin and beta2AR crystal structures into the electron microscopy map densities with constraints provided by HDX-MS and cross linking. This model, which was published in the June 22, 2014, edition of the journal Nature, provided valuable insights into the overall architecture of a receptor–arrestin complex.
“Arrestin’s primary role is to put the cap on GPCR signaling. Elucidating the structure of this complex is crucial for understanding how the receptors are desensitized in order to prevent aberrant signaling,” said co-senior author Dr. Georgios Skiniotis, professor of life sciences at the University of Michigan.
Related Links:
Duke University
University of Michigan
Stanford University