Kidney Transplant Rejection Linked to New Mismatch Locus
|
By LabMedica International staff writers Posted on 13 Jun 2019 |

Image: The NimbleGen MS 200 CGH LOH microarray detection scanner (Photo courtesy of Roche).
Approximately 20% of the kidney waiting list in the USA consists of candidates whose allografts have failed. Acute rejection is one of the strongest predictors of decreased allograft survival.
In the context of kidney transplantation, genomic incompatibilities between donor and recipient may lead to allosensitization against new antigens. A new form of genetic mismatch, or “genomic collision”, has been identified between kidney transplant recipients and organ donors that appears to increase the risk of allograft rejection.
A large team of scientists led by those at Columbia University (New York, NY, USA) performed a two-stage genetic association study of kidney allograft rejection. The study aimed at uncovering high-priority copy number variants influencing kidney allograft rejection in more than 700 transplant recipients. In particular, they focused on the so-called genomic collision situations, in which transplant recipients carried two copies of a deletion that was not homozygous in the organ donor.
The identification of alloantibodies was performed with the use of protein arrays, enzyme-linked immunosorbent assays (ELISAs), and Western blot analyses. The team used the NimbleGen array-based comparative genome hybridization to search for 50 suspicious deletions affecting protein-coding parts of the genome in 705 kidney transplant recipients. From there, they searched for associations with kidney allograft rejection in a “time-to-event survival analysis” in the patients, who were followed for more than eight-and-a-half years, on average.
In the discovery cohort, which included 705 recipients, they found a significant association with allograft rejection at the LIMS1 locus represented by rs893403 (hazard ratio with the risk genotype versus the non-risk genotypes, 1.84). This effect was replicated under the genomic-collision model in three independent cohorts involving a total of 2,004 donor–recipient pairs (hazard ratio, 1.55). In the combined analysis (discovery cohort plus replication cohorts), the risk genotype was associated with a higher risk of rejection than the non-risk genotype (hazard ratio, 1.63). The team identified a specific antibody response against LIMS1, a kidney-expressed protein encoded within the collision locus. The response involved predominantly IgG2 and IgG3 antibody subclasses.
The team got further evidence for the hypothesis following a functional analysis of the rejection-related risk variant, and sero-reactivity experiments that relied on protein array data for hundreds more kidney transplant recipients who did or did not experience allograft rejection. The authors noted that the LIMS1 protein also appears to be expressed in other commonly transplanted organs, including the heart and lung, though they cautioned that follow-up studies will be useful in determining whether our findings are generalizable to other organs. The study was published on May 16, 2019, in The New England Journal of Medicine.
Related Links:
Columbia University
In the context of kidney transplantation, genomic incompatibilities between donor and recipient may lead to allosensitization against new antigens. A new form of genetic mismatch, or “genomic collision”, has been identified between kidney transplant recipients and organ donors that appears to increase the risk of allograft rejection.
A large team of scientists led by those at Columbia University (New York, NY, USA) performed a two-stage genetic association study of kidney allograft rejection. The study aimed at uncovering high-priority copy number variants influencing kidney allograft rejection in more than 700 transplant recipients. In particular, they focused on the so-called genomic collision situations, in which transplant recipients carried two copies of a deletion that was not homozygous in the organ donor.
The identification of alloantibodies was performed with the use of protein arrays, enzyme-linked immunosorbent assays (ELISAs), and Western blot analyses. The team used the NimbleGen array-based comparative genome hybridization to search for 50 suspicious deletions affecting protein-coding parts of the genome in 705 kidney transplant recipients. From there, they searched for associations with kidney allograft rejection in a “time-to-event survival analysis” in the patients, who were followed for more than eight-and-a-half years, on average.
In the discovery cohort, which included 705 recipients, they found a significant association with allograft rejection at the LIMS1 locus represented by rs893403 (hazard ratio with the risk genotype versus the non-risk genotypes, 1.84). This effect was replicated under the genomic-collision model in three independent cohorts involving a total of 2,004 donor–recipient pairs (hazard ratio, 1.55). In the combined analysis (discovery cohort plus replication cohorts), the risk genotype was associated with a higher risk of rejection than the non-risk genotype (hazard ratio, 1.63). The team identified a specific antibody response against LIMS1, a kidney-expressed protein encoded within the collision locus. The response involved predominantly IgG2 and IgG3 antibody subclasses.
The team got further evidence for the hypothesis following a functional analysis of the rejection-related risk variant, and sero-reactivity experiments that relied on protein array data for hundreds more kidney transplant recipients who did or did not experience allograft rejection. The authors noted that the LIMS1 protein also appears to be expressed in other commonly transplanted organs, including the heart and lung, though they cautioned that follow-up studies will be useful in determining whether our findings are generalizable to other organs. The study was published on May 16, 2019, in The New England Journal of Medicine.
Related Links:
Columbia University
Latest Molecular Diagnostics News
- Blood Proteins Could Warn of Cancer Seven Years before Diagnosis
- New DNA Origami Technique to Advance Disease Diagnosis
- Ultrasound-Aided Blood Testing Detects Cancer Biomarkers from Cells
- New Respiratory Syndromic Testing Panel Provides Fast and Accurate Results
- New Synthetic Biomarker Technology Differentiates Between Prior Zika and Dengue Infections
- Novel Biomarkers to Improve Diagnosis of Renal Cell Carcinoma Subtypes
- RNA-Powered Molecular Test to Help Combat Early-Age Onset Colorectal Cancer
- Advanced Blood Test to Spot Alzheimer's Before Progression to Dementia
- Multi-Omic Noninvasive Urine-Based DNA Test to Improve Bladder Cancer Detection
- First of Its Kind NGS Assay for Precise Detection of BCR::ABL1 Fusion Gene to Enable Personalized Leukemia Treatment
- Urine Test to Revolutionize Lyme Disease Testing
- Simple Blood Test Could Enable First Quantitative Assessments for Future Cerebrovascular Disease
- New Genetic Testing Procedure Combined With Ultrasound Detects High Cardiovascular Risk
- Blood Samples Enhance B-Cell Lymphoma Diagnostics and Prognosis
- Blood Test Predicts Knee Osteoarthritis Eight Years Before Signs Appears On X-Rays
- Blood Test Accurately Predicts Lung Cancer Risk and Reduces Need for Scans
Channels
Clinical Chemistry
view channel
3D Printed Point-Of-Care Mass Spectrometer Outperforms State-Of-The-Art Models
Mass spectrometry is a precise technique for identifying the chemical components of a sample and has significant potential for monitoring chronic illness health states, such as measuring hormone levels... Read more.jpg)
POC Biomedical Test Spins Water Droplet Using Sound Waves for Cancer Detection
Exosomes, tiny cellular bioparticles carrying a specific set of proteins, lipids, and genetic materials, play a crucial role in cell communication and hold promise for non-invasive diagnostics.... Read more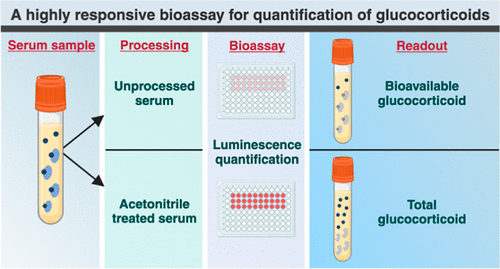
Highly Reliable Cell-Based Assay Enables Accurate Diagnosis of Endocrine Diseases
The conventional methods for measuring free cortisol, the body's stress hormone, from blood or saliva are quite demanding and require sample processing. The most common method, therefore, involves collecting... Read moreHematology
view channel
Next Generation Instrument Screens for Hemoglobin Disorders in Newborns
Hemoglobinopathies, the most widespread inherited conditions globally, affect about 7% of the population as carriers, with 2.7% of newborns being born with these conditions. The spectrum of clinical manifestations... Read more
First 4-in-1 Nucleic Acid Test for Arbovirus Screening to Reduce Risk of Transfusion-Transmitted Infections
Arboviruses represent an emerging global health threat, exacerbated by climate change and increased international travel that is facilitating their spread across new regions. Chikungunya, dengue, West... Read more
POC Finger-Prick Blood Test Determines Risk of Neutropenic Sepsis in Patients Undergoing Chemotherapy
Neutropenia, a decrease in neutrophils (a type of white blood cell crucial for fighting infections), is a frequent side effect of certain cancer treatments. This condition elevates the risk of infections,... Read more
First Affordable and Rapid Test for Beta Thalassemia Demonstrates 99% Diagnostic Accuracy
Hemoglobin disorders rank as some of the most prevalent monogenic diseases globally. Among various hemoglobin disorders, beta thalassemia, a hereditary blood disorder, affects about 1.5% of the world's... Read moreImmunology
view channel.jpg)
AI Predicts Tumor-Killing Cells with High Accuracy
Cellular immunotherapy involves extracting immune cells from a patient's tumor, potentially enhancing their cancer-fighting capabilities through engineering, and then expanding and reintroducing them into the body.... Read more
Diagnostic Blood Test for Cellular Rejection after Organ Transplant Could Replace Surgical Biopsies
Transplanted organs constantly face the risk of being rejected by the recipient's immune system which differentiates self from non-self using T cells and B cells. T cells are commonly associated with acute... Read more
AI Tool Precisely Matches Cancer Drugs to Patients Using Information from Each Tumor Cell
Current strategies for matching cancer patients with specific treatments often depend on bulk sequencing of tumor DNA and RNA, which provides an average profile from all cells within a tumor sample.... Read more
Genetic Testing Combined With Personalized Drug Screening On Tumor Samples to Revolutionize Cancer Treatment
Cancer treatment typically adheres to a standard of care—established, statistically validated regimens that are effective for the majority of patients. However, the disease’s inherent variability means... Read moreMicrobiology
view channel
Integrated Solution Ushers New Era of Automated Tuberculosis Testing
Tuberculosis (TB) is responsible for 1.3 million deaths every year, positioning it as one of the top killers globally due to a single infectious agent. In 2022, around 10.6 million people were diagnosed... Read more
Automated Sepsis Test System Enables Rapid Diagnosis for Patients with Severe Bloodstream Infections
Sepsis affects up to 50 million people globally each year, with bacteraemia, formerly known as blood poisoning, being a major cause. In the United States alone, approximately two million individuals are... Read moreEnhanced Rapid Syndromic Molecular Diagnostic Solution Detects Broad Range of Infectious Diseases
GenMark Diagnostics (Carlsbad, CA, USA), a member of the Roche Group (Basel, Switzerland), has rebranded its ePlex® system as the cobas eplex system. This rebranding under the globally renowned cobas name... Read more
Clinical Decision Support Software a Game-Changer in Antimicrobial Resistance Battle
Antimicrobial resistance (AMR) is a serious global public health concern that claims millions of lives every year. It primarily results from the inappropriate and excessive use of antibiotics, which reduces... Read morePathology
view channelHyperspectral Dark-Field Microscopy Enables Rapid and Accurate Identification of Cancerous Tissues
Breast cancer remains a major cause of cancer-related mortality among women. Breast-conserving surgery (BCS), also known as lumpectomy, is the removal of the cancerous lump and a small margin of surrounding tissue.... Read more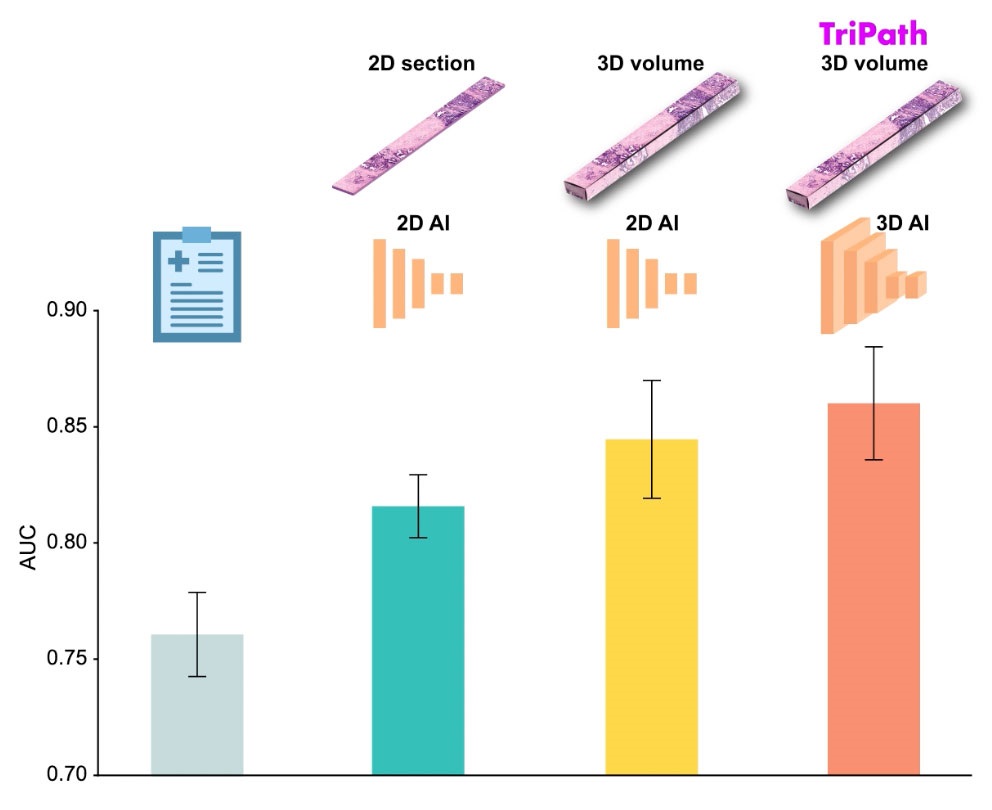
AI Advancements Enable Leap into 3D Pathology
Human tissue is complex, intricate, and naturally three-dimensional. However, the thin two-dimensional tissue slices commonly used by pathologists to diagnose diseases provide only a limited view of the... Read more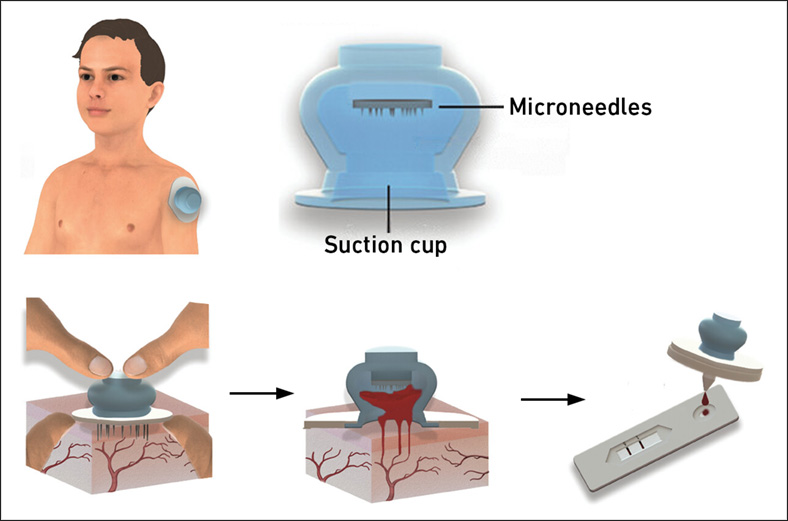
New Blood Test Device Modeled on Leeches to Help Diagnose Malaria
Many individuals have a fear of needles, making the experience of having blood drawn from their arm particularly distressing. An alternative method involves taking blood from the fingertip or earlobe,... Read more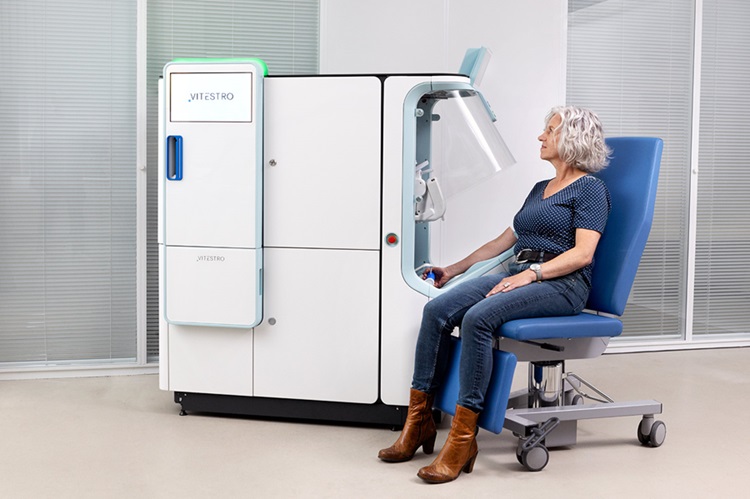
Robotic Blood Drawing Device to Revolutionize Sample Collection for Diagnostic Testing
Blood drawing is performed billions of times each year worldwide, playing a critical role in diagnostic procedures. Despite its importance, clinical laboratories are dealing with significant staff shortages,... Read moreTechnology
view channel
New Diagnostic System Achieves PCR Testing Accuracy
While PCR tests are the gold standard of accuracy for virology testing, they come with limitations such as complexity, the need for skilled lab operators, and longer result times. They also require complex... Read more
DNA Biosensor Enables Early Diagnosis of Cervical Cancer
Molybdenum disulfide (MoS2), recognized for its potential to form two-dimensional nanosheets like graphene, is a material that's increasingly catching the eye of the scientific community.... Read more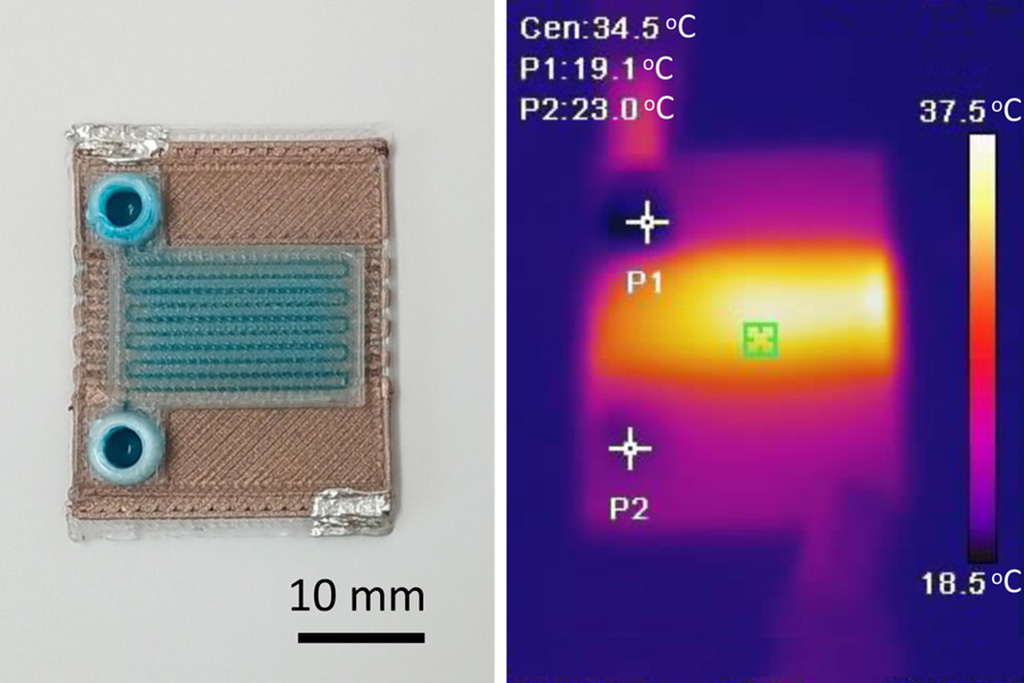
Self-Heating Microfluidic Devices Can Detect Diseases in Tiny Blood or Fluid Samples
Microfluidics, which are miniature devices that control the flow of liquids and facilitate chemical reactions, play a key role in disease detection from small samples of blood or other fluids.... Read more
Breakthrough in Diagnostic Technology Could Make On-The-Spot Testing Widely Accessible
Home testing gained significant importance during the COVID-19 pandemic, yet the availability of rapid tests is limited, and most of them can only drive one liquid across the strip, leading to continued... Read moreIndustry
view channel
Danaher and Johns Hopkins University Collaborate to Improve Neurological Diagnosis
Unlike severe traumatic brain injury (TBI), mild TBI often does not show clear correlations with abnormalities detected through head computed tomography (CT) scans. Consequently, there is a pressing need... Read more
Beckman Coulter and MeMed Expand Host Immune Response Diagnostics Partnership
Beckman Coulter Diagnostics (Brea, CA, USA) and MeMed BV (Haifa, Israel) have expanded their host immune response diagnostics partnership. Beckman Coulter is now an authorized distributor of the MeMed... Read more_1.jpg)