AI Tool Accurately Determines Breast Cancer Prognosis
Posted on 19 Jul 2025
A new study has found that cells and tissues surrounding a breast cancer tumor may hold critical information about how patients will respond to treatment. The research, published in the journal Patterns, marks a significant step toward developing artificial intelligence (AI) tools to help oncologists more accurately determine prognoses and decide which treatments may be most effective on a case-by-case basis.
Researchers at Johns Hopkins University (Baltimore, MD, USA) have designed an interpretable machine learning model that analyzed images of microscopic tissues and biopsy samples from 579 breast cancer patients who were undergoing standard treatments such as chemotherapy. The model examined both the tumor and the surrounding tissue, assessing every cell’s position in relation to the tumor and interactions with neighboring cells. This allowed the algorithm to detect 66 unique cell patterns and categorize patients into seven prognostic groups based on these patterns. Unlike conventional AI methods, the model could clearly identify which specific elements contributed to each pattern, allowing researchers to reverse-engineer the data and uncover the driving factors behind the groupings.
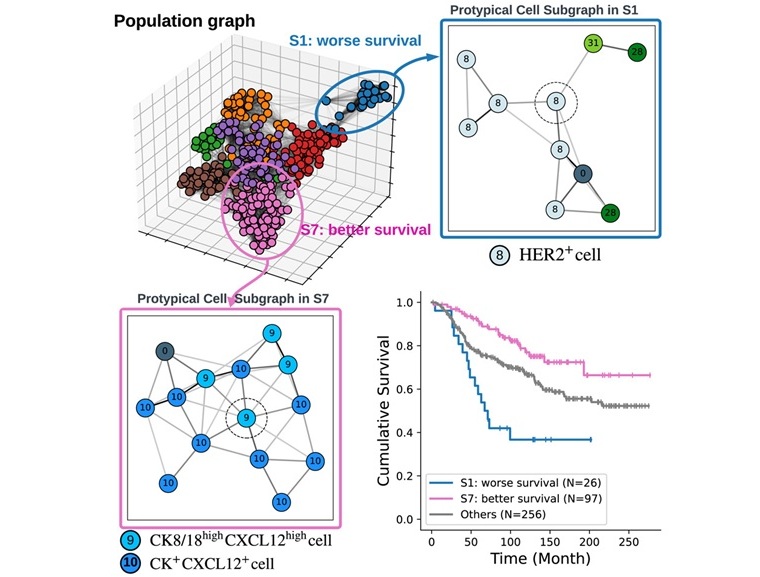
The model’s validation revealed several significant findings. One group of patients with the best survival outcomes had a distinct combination of three cell types: CK8-18high, CXCL12high, and CK+CXCL12+. On the other hand, patients with self-aggregated HER2+ tumor cells had the worst outcomes—a result that aligns with the known aggressiveness of HER2-positive breast cancers. Additionally, triple-negative breast cancer patients with well-organized immune cells surrounding their tumors also had better prognoses than others with the same condition. These results suggest that analyzing spatial tissue patterns may provide oncologists with valuable biomarkers for predicting patient outcomes and personalizing treatment strategies. Moving forward, the researchers plan to expand their methodology to other imaging technologies and apply it to different types of cancer to uncover more actionable insights.
"We're developing a toolbox that you can use anytime you have a massive data set and you don't want to start with an explicit hypothesis," said first author Zhenzhen Wang, a PhD candidate in biomedical engineering at Johns Hopkins University. "Typically, you'd have to design a study to ask if one specific pattern of cells is important, and check if the answer is yes or no. But with our model, we can save time by asking open-ended questions about which patterns are important when trying to understand how patients will respond to treatment or have a better chance of survival."
Related Links:
Johns Hopkins University