Malignant Pleural Mesothelioma Personalized by ctDNA Profiling
By LabMedica International staff writers
Posted on 09 Apr 2019
Malignant pleural mesothelioma (MPM) is an aggressive tumor associated with exposure to asbestos and is rising in incidence worldwide. A lack of early detection methods and treatment successes has maintained a bleak prognosis of just 12 to 16 months.Posted on 09 Apr 2019
Circulating tumor DNA (ctDNA) is emerging as an important stratification biomarker in other thoracic malignancies, however studies in this area has been lacking in MPM. Homozygous deletion of the cyclin-dependent kinase inhibitor 2A gene (CDKN2A) is one of the few established negatively prognostic molecular markers in MPM.
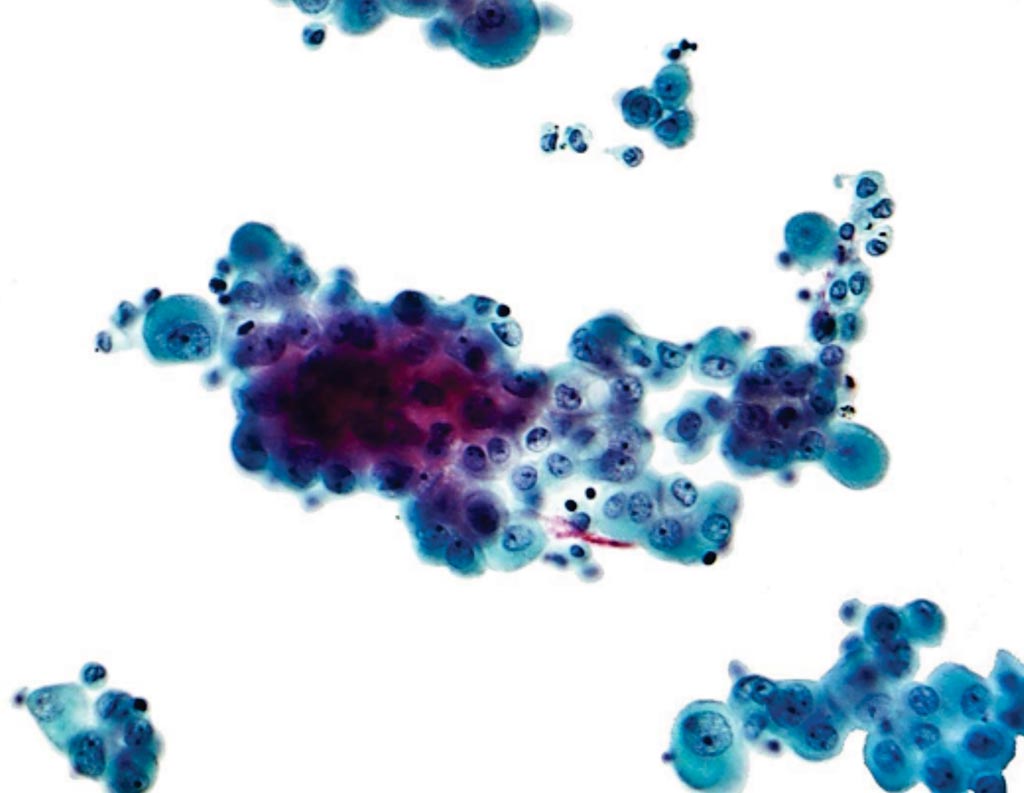
Image: A photomicrograph of a pleural fluid cytopathology specimen showing mesothelioma (Photo courtesy of Nephron).
Scientists at the University of Leicester (Leicester, UK) and their colleagues conducted multiregional whole exome sequencing (m-WES) on tumor tissue (four or five regions) and matched germline DNA from 11 patients with MPM that received surgery (extended pleurectomy decortication) as their primary treatment modality. The team used a validated bioinformatics pipeline (developed as part of the TRACERx study), clonal single nucleotide variants (SNVs) and/or indels (present within all tumor regions) were selected from m-WES data for patient-specific assay design. Variants were prioritized for selection based on tumor driver status and higher mean variant allele frequencies (VAFs) amongst tumor regions. A minimum of 10 ng cfDNA isolated from pre-surgical blood was analyzed for ctDNA detection using droplet digital PCR (ddPCR), with positivity being defined as ≥ 3 mutant droplets and a plasma VAF ≥ 0.1%.
The investigators reported that 4/11 (36.4%) patients were ctDNA-positive using this patient-specific approach with VAFs ranging between 0.16% and 2.96%. Two additional patients had one or two positive mutant droplets, but these were excluded as potential false positives. In this pilot study, ctDNA-positive patients had significantly shorter survival than ctDNA-negative patients, although this failed to reach significance by Cox regression analysis, likely due to a small sample size. Furthermore, in this cohort ctDNA status was a stronger prognostic biomarker than CDKN2A deletion status.
The authors conclude that considering the short survival of patients with MPM despite surgery, they considered ctDNA-based analyses could be used to stratify patients regarding surgery, where risk-benefit would be marginal for those ctDNA-positives. This study provides proof-of-principle that the detection of tumor DNA from minimally invasive sources is possible in MPM and has alluded to how this could potentially impact clinical decision-making. The study was presented at the American Association of Cancer Research Annual Meeting, held March 29 to Apr 3, 2019, in Atlanta, GA, USA.
Related Links:
University of Leicester