Panel of Long Noncoding RNAs Predicts Prospects for Leukemia Survival
By LabMedica International staff writers
Posted on 22 Dec 2014
Cancer researchers have identified a panel of 48 long noncoding RNAs that is indicative of a favorable outcome in older patients suffering from acute myeloid leukemia (AML) and has the potential to aid clinicians in choosing the least toxic, most effective treatment for this disease, which has a three-year survival rate of only 5% to 15%.Posted on 22 Dec 2014
Long non-coding RNAs (long ncRNAs, lncRNA) are non-protein coding transcripts longer than 200 nucleotides. This somewhat arbitrary limit distinguishes long ncRNAs from small regulatory RNAs such as microRNAs (miRNAs, short interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and other short RNAs. While lncRNAs are known to be involved in numerous biological roles including imprinting, epigenetic gene regulation, cell cycle and apoptosis, and metastasis and prognosis in solid tumors, their role in AML has not been clarified.
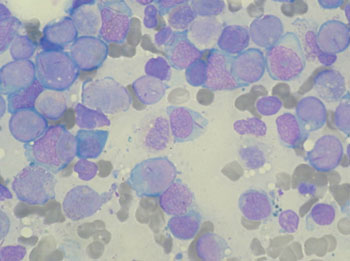
Image: Bone marrow aspirate showing acute myeloid leukemia (Photo courtesy of Wikimedia Commons).
To determine whether lncRNAs are associated with clinical features and recurrent mutations in older patients (aged over 60 years) with cytogenetically normal (CN)-AML, investigators at Ohio State University (Columbus, USA) evaluated lncRNA expression in 148 untreated older CN-AML patients using a custom microarray platform. An independent set of 71 untreated older patients with CN-AML was used to validate the outcome scores using RNA sequencing.
The investigators derived an lncRNA score comprising 48 lncRNAs. Patients with an unfavorable compared with favorable lncRNA score had a lower complete response rate and shorter disease-free survival. Three years into the study, only 7% of patients with an unfavorable score were disease free compared with 39% of patients with a favorable score. Overall survival at three years for those with an unfavorable score was 10% versus 43% for patients with a favorable score.
“We have identified a pattern of 48 lncRNAs that predicted both response to standard chemotherapy and overall survival in older CN-AML patients,” said first author Dr. Ramiro Garzon, associate professor of internal medicine at Ohio State University.
“Patients in the favorable group had a high probability of responding to standard chemotherapy, while those in the unfavorable group generally responded poorly to the treatment and had worse overall survival.”
The study was published in the December 15, 2014, online edition of the journal Proceedings of the National Academy of Sciences of the United States of America (PNAS).
Related Links:
Ohio State University