Chronic Lymphocytic Leukemia Genomic Study Reveals New Subtypes
Posted on 09 Aug 2022
Chronic lymphocytic leukemia (CLL) is a type of cancer that affects the blood and bone marrow. It affects the white blood cells called lymphocytes. It tends to develop very slowly. It is more common in older people and is rare in people younger than 40.
Many people with CLL are asymptomatic and are diagnosed because they have a routine blood test for something else. In CLL symptoms tend to be mild at first and get worse slowly. Many symptoms are vague Recent advances in cancer characterization have consistently revealed marked heterogeneity, impeding the completion of integrated molecular and clinical maps for each malignancy.
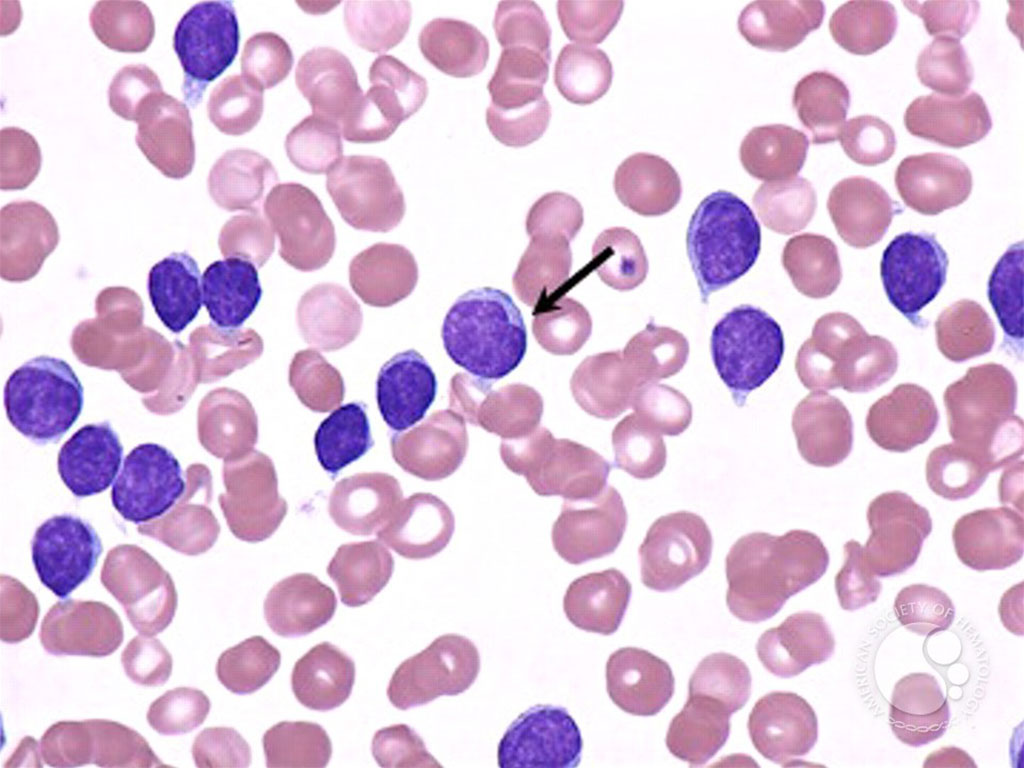
An international team of medical scientists, led by those at the Massachusetts General Cancer Center (Boston, MA, USA), have characterized the somatic mutation, gene expression, and epigenetic changes that occur within and across CLL subtypes, uncovering alterations with potential prognostic impact. CLL is a B cell neoplasm with variable natural history that is conventionally categorized into two subtypes distinguished by extent of somatic mutations in the heavy-chain variable region of immunoglobulin genes (IGHV).
The team assessed mutation, structural variant, gene expression, and regulatory features in pre-treatment, post-treatment, or treatment refractory/relapsed tumor samples from 1,148 individuals with CLL or monoclonal B cell lymphocytosis. Nearly 1,100 of the samples were subjected to exome or whole-genome sequencing, while 712 were assessed with RNA sequencing, and DNA methylation profiling was done on 999 samples.
The investigator’s results highlighted 202 suspected driver genes, impacted by recurrent alterations ranging from single nucleotide changes or small insertions or deletions to structural variants and DNA methylation shifts that appeared to drive CLL, including 109 suspected driver mutations not linked to the blood and bone marrow malignancy in the past. The scientists identified copy number mutations or structural variants that distinguished CLL subtypes with or without mutations in the immunoglobulin gene heavy-chain variable region (IGHV) through an analysis of 512 mutated IGHV (M-CLL) and 459 IGHV-unmutated CLL tumors.
The team noted that specific driver gene changes within IGHV subtypes appeared to correspond to clinical outcomes in CLL, as did drivers found through mutation, structural variant, gene expression, or methylation analyses, though combining the different data streams appeared to offer the most complete view of the disease. The investigators are making the new CLL omics data available to other scientists, along with corresponding clinical data, in an effort to unearth still other insights that may improve the understanding, management, or treatment of CLL.
Catherine Wu, MD, a professor of medicine and co-senior and co-corresponding author, said, “Such a CLL map could eventually be leveraged in the clinic, wherein the genomic features of new patients can be compared with the treatments and outcomes of patients with similar genetic profiles. This profiling could potentially help more accurately tailor prognosis and treatment of a new patient based on their particular molecular features, getting closer to precision medicine.” The study was published on August 4, 2022 in the journal Nature Genetics.
Related Links:
Massachusetts General Cancer Center