Biomarkers Could Give Cancer Patients Better Survival Estimates
By LabMedica International staff writers
Posted on 21 Jun 2016
Cancer patients are often told by their doctors approximately how long they have to live, and how well they will respond to treatments, but there is a way to improve the accuracy of doctors' predictions.Posted on 21 Jun 2016
A new method has been developed that could eventually lead to a way to do just that, using data about patients' genetic sequences to produce more reliable projections for survival time and how they might respond to possible treatments.
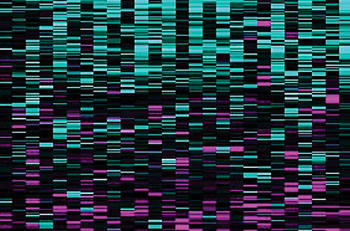
Image: A SURVIV analysis of breast cancer isoforms developed at UCLA. Blue lines are associated with longer survival times, and magenta lines with shorter survival times (Photo courtesy of Professor Yi Xing).
Scientists at the University of California-Los Angeles (UCLA, CA, USA) and their colleagues have developed a method that analyzes various gene isoforms using data from ribonucleic acid (RNA) molecules in cancer specimens. These isoforms are combinations of genetic sequences that can produce an enormous variety of RNAs and proteins from a single gene.
That process, called RNA sequencing, or RNA-seq, reveals the presence and quantity of RNA molecules in a biological sample. In the method developed, the scientists analyzed the ratios of slightly different genetic sequences within the isoforms, enabling them to detect important but subtle differences in the genetic sequences. In contrast, the conventional analysis aggregates all of the isoforms together, meaning that the technique misses important differences within the isoforms.
The scientists studied tissues from 2,684 people with cancer whose samples were part of the National Institutes of Health's Cancer Genome Atlas, and they spent more than two years developing the algorithm for SURVIV (for "survival analysis of mRNA isoform variation"). The team has identified some 200 isoforms that are associated with survival time for people with breast cancer; some predict longer survival times, others are linked to shorter times. Armed with that knowledge, the scientists might eventually be able to target the isoforms associated with shorter survival times in order to suppress them and fight disease. They evaluated the performance of survival predictors using a metric called C-index and found that across the six different types of cancer they analyzed, their isoform-based predictions performed consistently better than the conventional gene-based predictions.
Yi Xing, PhD, an assistant professor and senior author of the study, said, “Our finding suggests that isoform ratios provide a more robust molecular signature of cancer patients in large-scale RNA-seq datasets. In cancer, sometimes a single gene produces two isoforms, one of which promotes metastasis and one of which represses metastasis.” The study was published on June 9, 2016, in the journal Nature Communications.
Related Links:
University of California-Los Angeles