CRISPR-Based Microarray System Enables Massive Testing for Viral Pathogens
By LabMedica International staff writers
Posted on 11 May 2020
A novel microfluidic nucleic acid-based diagnostic system is capable of detecting a specific virus from a catalog of more than 160 human pathogens, including the COVID-19 coronavirus, simultaneously in more than 1,000 samples.Posted on 11 May 2020
To enable routine surveillance and comprehensive diagnostic applications, there is a need for detection technologies that can be scaled-up to test many samples while simultaneously testing for multiple individual pathogens. To accomplish this, investigators at the Broad Institute of MIT and Harvard (Cambridge, MA, USA) developed a diagnostic platform called CARMEN (Combinatorial Arrayed Reactions for Multiplexed Evaluation of Nucleic acids). The CARMEN system depends on nanoliter droplets containing CRISPR/Cas 13-based nucleic acid detection reagents.
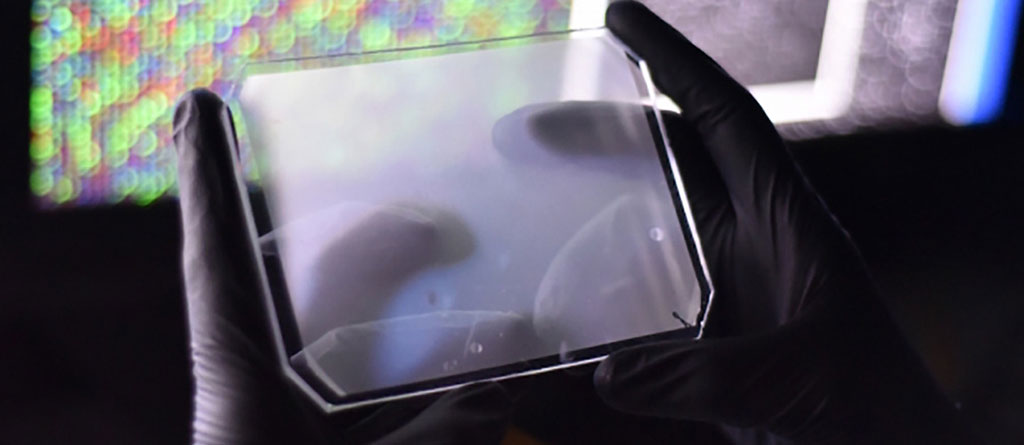
Image: Photograph of the CARMEN microwell chip (Photo courtesy of Michael James Butts)
CRISPRs (clustered regularly interspaced short palindromic repeats) are segments of prokaryotic DNA containing short repetitions of base sequences. Each repetition is followed by short segments of "spacer DNA" from previous exposures to a bacterial virus or plasmid.
Recent computational efforts to identify new CRISPR systems uncovered a novel type of RNA targeting enzyme, Cas13. The diverse Cas13 family contains at least four known subtypes, including Cas13a (formerly C2c2), Cas13b, Cas13c, and Cas13d. Cas13a was shown to bind and cleave RNA, protecting bacteria from RNA phages and serving as a powerful platform for RNA manipulation. It was suggested that Cas13a could function as part of a versatile, RNA-guided RNA-targeting CRISPR/Cas system holding great potential for precise, robust, and scalable RNA-guided RNA-targeting applications.
The CARMEN platform comprises a rubber chip, slightly larger than a smartphone, containing tens of thousands of microwells designed to each hold a pair of nanoliter-sized droplets. One droplet contains viral genetic material from a sample, and the other contains virus-detection reagents.
Detection of viral nucleic acids is by a modification of the SHERLOCK protocol. This is a method for single molecule detection of nucleic acid targets and stands for Specific High Sensitivity Enzymatic Reporter unLOCKing. It works by amplifying genetic sequences and programming a CRISPR molecule to detect the presence of a specific genetic signature in a sample, which can also be quantified. When it finds those signatures, the CRISPR enzyme is activated and releases a robust signal. This signal can be adapted to work on a simple paper strip test, in laboratory equipment, or to provide an electrochemical readout that can be read with a mobile phone.
In the CARMEN platform, nanoliter droplets containing CRISPR-based nucleic acid detection reagents self-organized in the microwell array to pair with droplets of amplified samples, testing each sample against each CRISPR RNA (crRNA) in replicate. The combination of CARMEN and Cas13 detection (CARMEN-Cas13) enabled robust testing of more than 4,500 crRNA-target pairs on a single array. The entire protocol, from RNA extraction to results, required less than eight hours.
Employing the CARMEN-Cas13 method, the investigators developed a multiplexed assay that simultaneously differentiated 169 human-associated viruses with more than 10 published genome sequences and rapidly incorporated an additional crRNA to detect the causative agent of the COVID-19 pandemic coronavirus. CARMEN-Cas13 further enabled comprehensive subtyping of influenza A strains and multiplexed identification of dozens of HIV drug-resistance mutations.
"This miniaturized approach to diagnostics is resource-efficient and easy to implement," said co-senior author Dr. Paul Blainey, associate professor of biological engineering at the Massachusetts Institute of Technology. "New tools require creativity and innovation, and with these advances in chemistry and microfluidics, we are enthusiastic about the potential for CARMEN as the community works to beat back both COVID-19 and future infectious disease threats."
The CARMEN-Cas13 method was described in the April 29, 2020, online edition of the journal Nature.
Related Links:
Broad Institute of MIT and Harvard