Dual Nucleic Acid Template Assay Developed for Cancer
By LabMedica International staff writers
Posted on 16 Jan 2020
Massive parallel sequencing or massively parallel sequencing is any of several high-throughput approaches to DNA sequencing using the concept of massively parallel processing; it is also called next-generation sequencing or second-generation sequencing.Posted on 16 Jan 2020
Targeted next-generation sequencing is a powerful method to comprehensively identify biomarkers for cancer. Starting material is currently either DNA or RNA for different variations, but splitting to two assays is burdensome and sometimes unpractical, causing delay or complete lack of detection of critical events, in particular, potent and targetable fusion events.
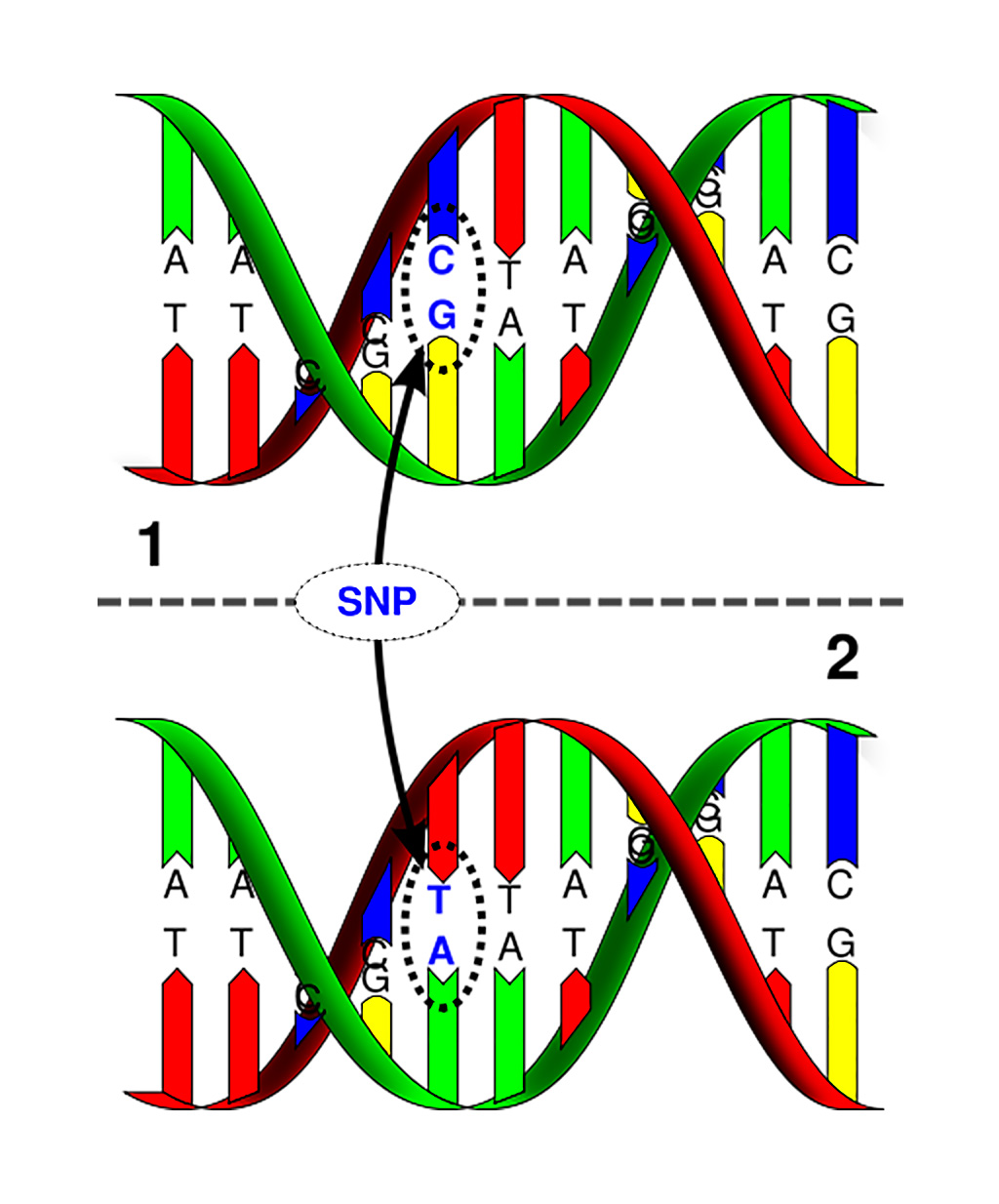
Image: Single nucleotide polymorphism: The upper DNA molecule differs from the lower DNA molecule at a single base-pair location (Photo courtesy of Marshall University).
Scientists at Zhejiang Cancer Hospital (Hangzhou, China) and their associates developed a single-tube, dual-template assay and an integrated bioinformatics pipeline for relevant variant calling. RNA was used for fusion detection, whereas DNA was used for single-nucleotide variations (SNVs) and insertion and deletions (indels). The reaction chemistry featured barcoded adaptor ligation, multiplexed linear amplification, and multiplexed polymerase chain reaction (PCR) for noise reduction and novel fusion detection. An auxiliary quality control assay was also developed.
The investigators reported that in a 1,000-sample lung tumor cohort, they identified all major SNV/indel hotspots and fusions, as well as MET exon 14 skipping and several novel or rare fusions. The occurrence frequencies were in line with previous reports and were verified by Sanger sequencing. One noteworthy fusion event was HLA-DRB1-MET that constituted the second intergenic MET fusion ever detected in lung cancer.
The authors concluded that their method should benefit not only a majority of patients carrying core actionable targets but also those with rare variations. Future extension of this assay to RNA expression and DNA copy number profiling of target genes such as programmed death-ligand 1 may provide additional biomarkers for immune checkpoint therapies. The study was published on the December 2019 issue of the journal Clinical Chemistry.
Related Links:
Zhejiang Cancer Hospital