Protein Atlas Accelerates Personalized Medicine in Leukemia Patients
By LabMedica International staff writers
Posted on 30 Apr 2019
Acute myelogenous leukemia is associated with risk factors that are largely unknown and with a heterogeneous response to treatment. Only about one in four people diagnosed with acute myelogenous leukemia (AML) survive five years after the initial diagnosis.Posted on 30 Apr 2019
To improve that survival rate, scientists have created an online atlas to identify and classify protein signatures present at AML diagnosis. The new protein classifications will help clinicians recommend better treatment and personalized medicine for patients suffering from this aggressive cancer, which occurs in the blood and bone marrow.
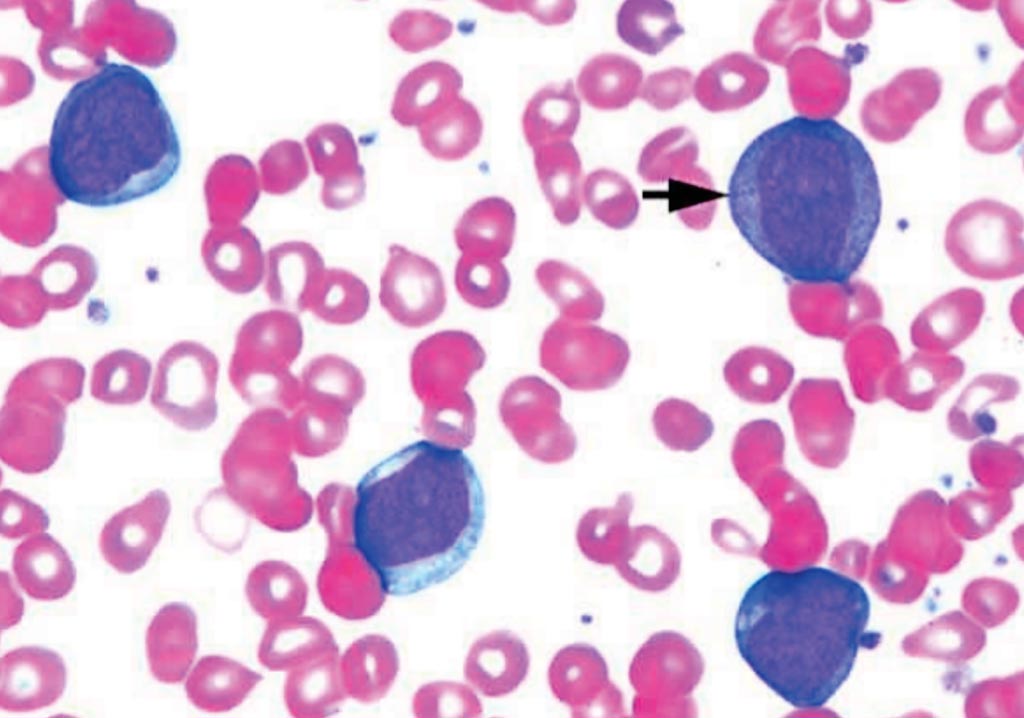
Image: Blood film of a patient with acute myelogenous leukemia defined by presence of more than 90% myeloblasts in blood and/or bone marrow (Photo courtesy of Pathpedia).
A team of scientists at the University of Texas at San Antonio (UTSA, San Antonio, TX, USA) and the University of Texas MD Anderson Cancer Center (Houston, TX, USA) examined the genetic, epigenetic and environmental diversity that occurs in cancerous cells due to AML. They analyzed proteomic screens of 205 patient biopsies and developed a new computational method called MetaGalaxy to categorize the protein signatures into 154 different patterns based on their cellular functions and pathways.
By approaching this challenge through the unique lens of developing a quantitative map for each leukemia patient from protein expression in their blood and bone marrow, rather than the standard lens of qualitative metrics and genetic risks alone, the collaborators will be able to more precisely categorize patients into risk groups and better predict their treatment outcomes. The team found 11 constellations of correlated functional patterns and 13 signatures that stratify the outcomes of patients. The scientists found limited overlap between proteomics data and both cytogenetics and genetic mutations. Moreover, leukemia cell lines show limited proteomic similarities with cells from patients with AML, suggesting that a deeper focus on patient-derived samples is needed to gain disease-relevant insights.
Amina Qutub, PhD, an associate professor and Biochemical Engineer and a senior study author said, “Acute myelogenous leukemia presents as a cancer so heterogeneous that it is often described as not one, but a collection of diseases. To decipher the clues found in proteins from blood and bone marrow of leukemia patients, we developed a new computer analysis, MetaGalaxy that identifies molecular hallmarks of leukemia. These hallmarks are analogous to the way constellations guide navigation of the stars: they provide a map to protein changes for leukemia.” The study was published on April 15, 2019, in the journal Nature Biomedical Engineering.
Related Links:
University of Texas at San Antonio
University of Texas MD Anderson Cancer Center