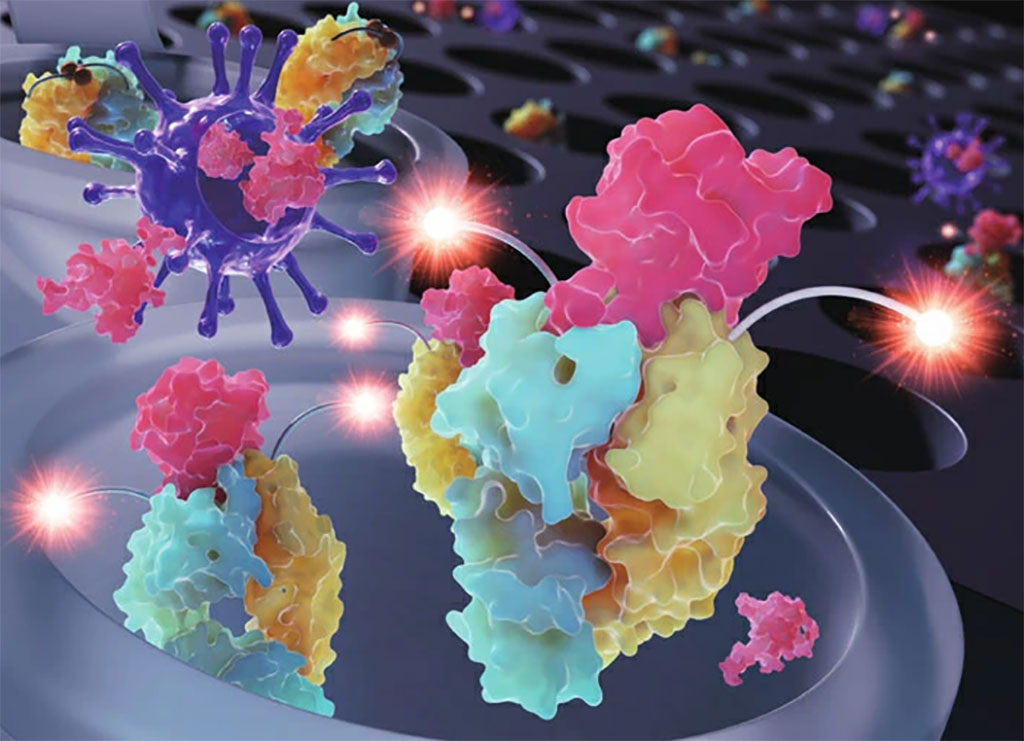
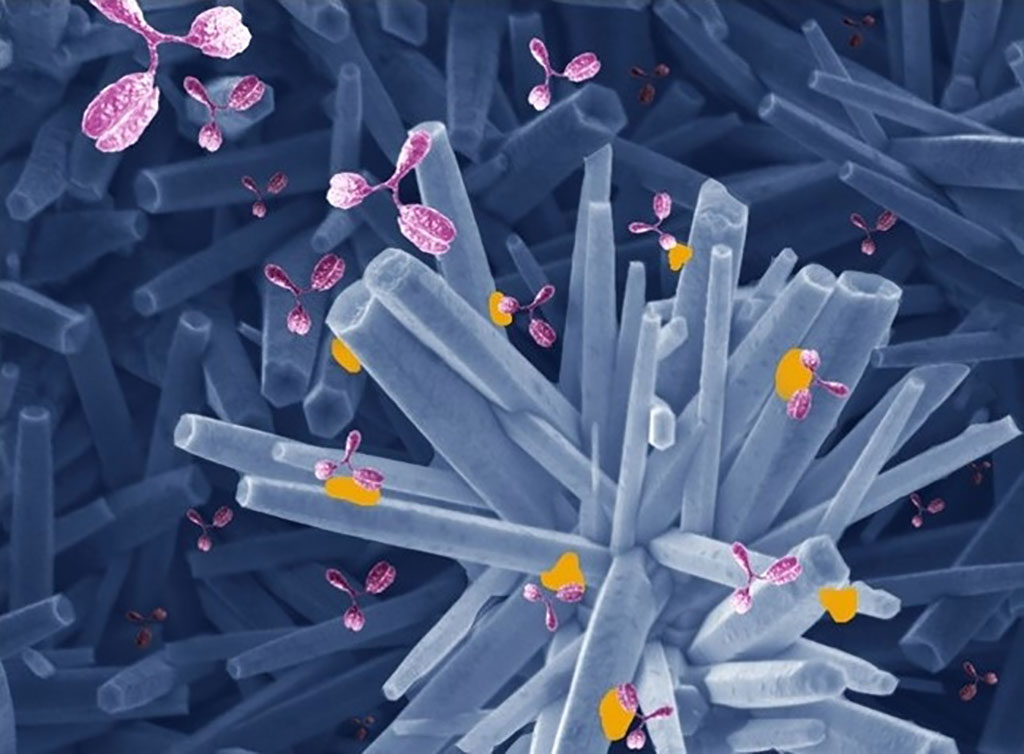
World-First Method for Rapid Isolation and Characterization of COVID-19 Variants
Posted on 01 Jun 2022
Researchers have developed a methodology that speeds up the process for isolating and characterizing the risks posed by COVID-19 variants of concern. The methodology, which has been shared with the global scientific community through publication in Nature Microbiology, is highly cost-effective, automated, and can be used to measure the effectiveness of therapeutics and scaled to test thousands of samples.
The methodology, called R-20, was developed by researchers from the Kirby Institute at UNSW Sydney (Sydney, Australia) and used to analyze all major circulating viral variants in 2021 that were identified from patients in hotel quarantine. At the end of 2021, it was applied to Australia’s first Omicron samples. The researchers found that the vaccine-induced antibody response to Omicron was 15-20-fold weaker than to the original SARS-CoV-2 strain. The rapid analysis included details of how well the variant evades antibodies and how resilient it is. The results have been cited in Centers for Disease Control and Prevention (CDC) guidelines, World Health Organization (WHO) technical reports and by the Australian Technical Advisory group on Immunization (ATAGI) regarding the use of booster vaccinations.
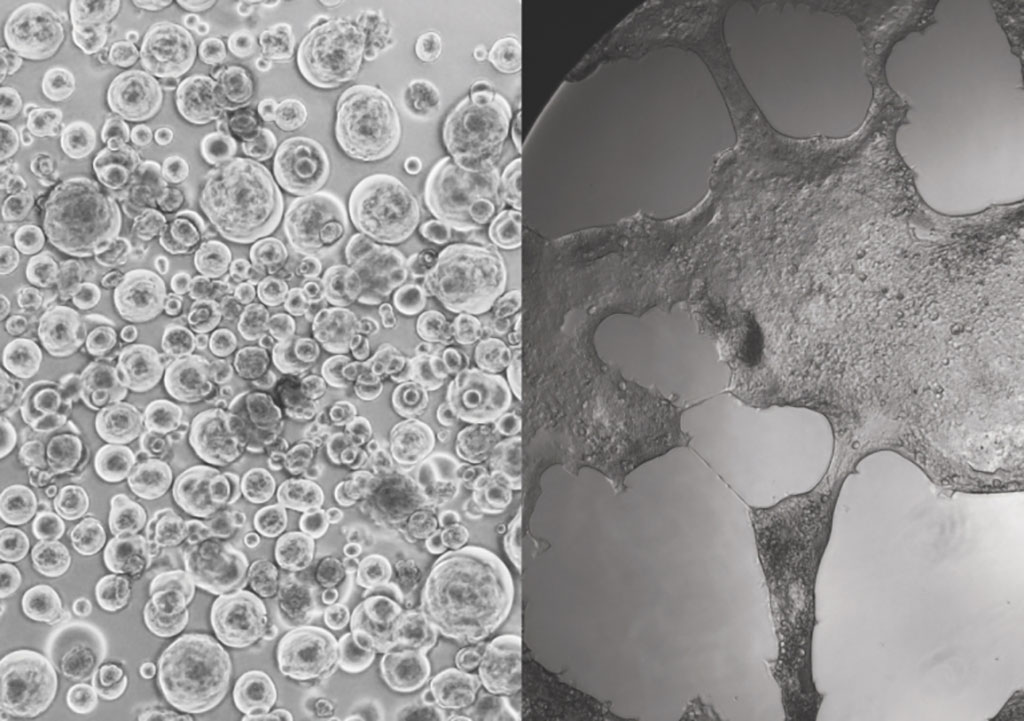
In order to develop the methodology, the researchers took lessons learned from HIV to develop cells with receptors that viruses ‘like’. They looked at hundreds of different cells to identify ones that would allow the virus to replicate as quickly and effectively as possible. They termed these cells as the ‘canaries in the coal mine’ – because they effectively die in the process of telling the team what each variant is up to. They also allow the team to capture variants of the virus quicker than any other cell culture method, with sensitivities approaching that of a PCR swab test.
Using R-20, the researchers look at viral properties of variants to understand how sick the virus is likely to make the population, as well as how evasive it is to existing vaccines and treatments. Identifying this quickly is crucial to inform public health policies such as vaccination strategies, which therapeutics will still work and advice on mask use to mitigate spread of variants to vulnerable populations such as the elderly.
“What makes our R-20 approach unique is its speed and accuracy,” says Associate Professor Turville who developed the methodology. “The best way to quickly understand how a virus works is to genetically develop a cell with receptors that the virus likes. R-20 uses 'supercharged' cells that allow the virus to replicate four times faster than through any other techniques currently published in the scientific literature.”
Related Links:
UNSW Sydney