Quick Ultra-Sensitive System Identifies Multidrug-Resistant Bacteria
By LabMedica International staff writers
Posted on 22 Dec 2020
The worldwide emergence of multidrug‐resistant (MDR) bacteria is associated with significant morbidity, mortality, and healthcare costs. Rapid and accurate diagnostic methods to detect antibiotic resistance are critical for antibiotic stewardship and infection control measurements.Posted on 22 Dec 2020
Traditional methods for detecting resistance are based on cultivating bacteria and testing their sensitivity to a spectrum of antibiotics. These methods take 48 to 72 hours to deliver results, and some strains of bacteria are difficult to cultivate. Molecular biological tests are a great deal faster, but even this method does not deliver satisfactory results for every bacterium.
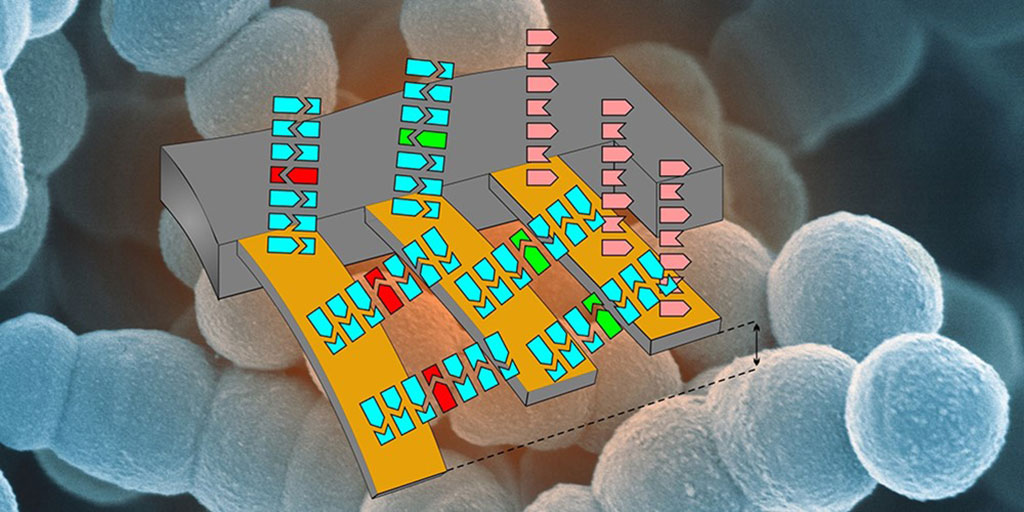
Image: Schematic illustration of the cantilever array to detect antibiotic resistance. (Photo courtesy of Department of Physics and Nano Imaging Lab, SNI, University of Basel).
A team of scientists from the Swiss Nanoscience Institute (SNI, Basel, Switzerland) have developed a cantilever testing system that allowed them to detect RNA from a single antibiotic resistant bacterium. The bacterial isolates, Pseudomonas aeruginosa and Enterococcus faecium, used were from the biobank of the division of Clinical Bacteriology and Mycology at the University Hospital Basel (Basel, Switzerland). Extracted RNA was quality controlled using Invitrogen Qubit 3.0 and NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA).
With the new cantilever system, it is not necessary to amplify or label the samples for analysis. The team began by attaching sequences of three genes associated with vancomycin resistance to the cantilevers and then exposed these prepared cantilevers to a flow of RNA extracted from bacteria. If RNA molecules from the resistance genes were present, the matching RNA fragments would bind to the cantilevers, causing them to undergo nanoscale deflection that could be detected using a laser.
The team found a clear signal even with point mutations. This method allowed the detection of not only resistance genes, but also individual point mutations associated with them. To study this, the scientists used point mutations coupled to genes responsible for resistance to ampicillin and other betalactam antibiotics.
François Huber, Dr phil nat, a professor and first author of the study, said, “The big advantage of the method we've developed is its speed and sensitivity. We succeeded in detecting tiny quantities of specific RNA fragments within five minutes.” In the case of single mutations, the detected RNA quantities corresponded to about 10 bacteria. When it came to detecting entire resistance genes, the investigators obtained a clear signal even with an amount of RNA that corresponded to a single bacterium. The study was published on November 30, 2020 in the journal Global Challenges.
Related Links:
Swiss Nanoscience Institute
University Hospital Basel
Thermo Fisher Scientific