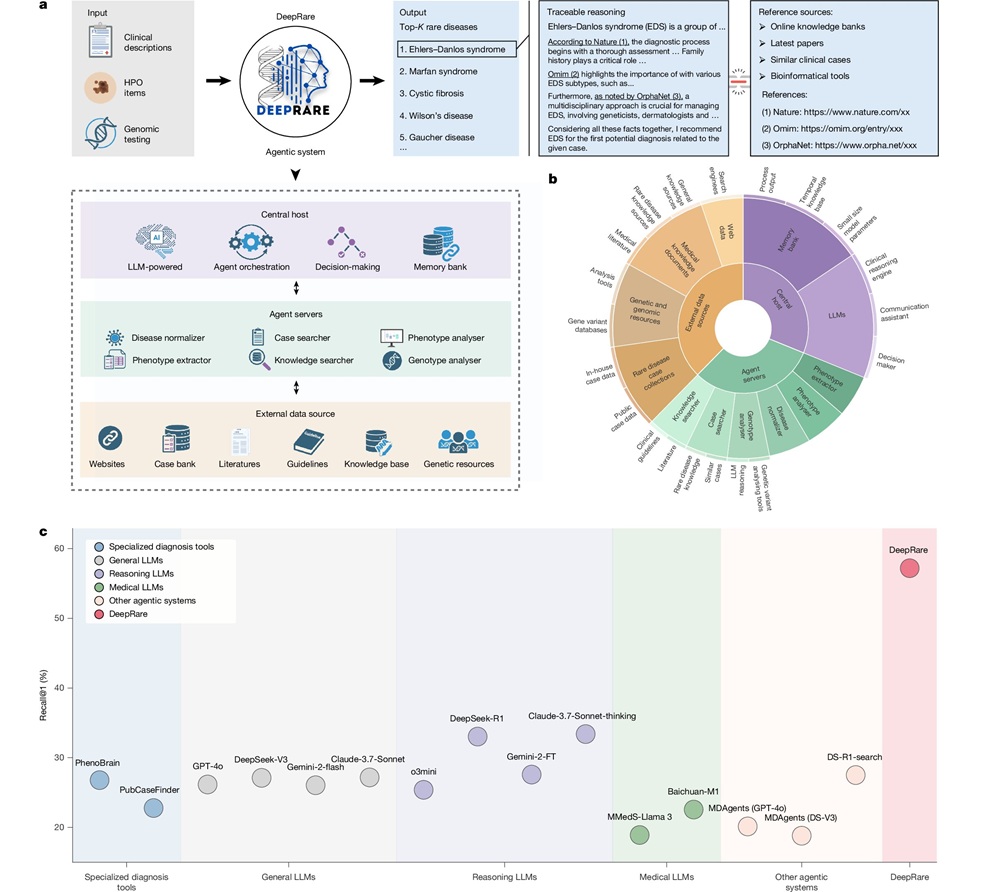
New Target Sequence Capture Panel Eases Detection of Viruses in Patient Samples
By LabMedica International staff writers
Posted on 15 Oct 2015
A recently developed targeted sequence capture panel, ViroCap, which was designed to enrich nucleic acid from DNA and RNA viruses that infect vertebrate hosts, allowed the rapid detection of unknown viruses in patient samples.Posted on 15 Oct 2015
Metagenomic shotgun sequencing (MSS) is an important tool for characterizing viral populations. It is culture-independent, requires no a priori knowledge of the viruses in the sample, and may provide useful genomic information. However, MSS can lack sensitivity and may yield insufficient data for detailed analysis. To solve these problems investigators at Washington University School of Medicine (St. Louis, MO, USA) have created a targeted sequence capture panel, ViroCap, designed to enrich nucleic acid from DNA and RNA viruses from 34 families that infect vertebrate hosts.
![Image: A new test can detect virtually any virus that infects people and animals, including the Ebolavirus depicted in this enhanced electron micrograph (Photo courtesy of the [US] National Institute of Allergy and Infectious Diseases). Image: A new test can detect virtually any virus that infects people and animals, including the Ebolavirus depicted in this enhanced electron micrograph (Photo courtesy of the [US] National Institute of Allergy and Infectious Diseases).](https://globetechcdn.com/mobile_labmedica/images/stories/articles/article_images/2015-10-14/GMS-353.jpg)
Image: A new test can detect virtually any virus that infects people and animals, including the Ebolavirus depicted in this enhanced electron micrograph (Photo courtesy of the [US] National Institute of Allergy and Infectious Diseases).
During the development of ViroCap the investigators submitted consolidated target sequences to 454 Roche NimbleGen (Madison, WI, USA) for capture library design and 454 synthesis. This computational approach condensed nearly one billion base pairs (bps) of viral reference sequence into less than 200 million bps of unique, representative sequence suitable for targeted sequence capture. These sequences were used as probes to scoop up viruses in patient samples that were a genetic match. The matched viral material was analyzed using high-throughput genetic sequencing.
The investigators used ViroCap to analyze two sets of samples, one derived from samples submitted to a diagnostic virology laboratory and one derived from samples collected in a study of fever in children. They reported detecting 14 and 18 viruses in the two sets, respectively, comprising 19 genera from 10 families. Compared to standard genomics-based detection methods, 52% more viruses were found by the ViroCap method.
"With this test, you do not have to know what you are looking for," said senior author, Dr. Gregory Storch, professor of pediatrics at Washington University School of Medicine. "It casts a broad net and can efficiently detect viruses that are present at very low levels. We think the test will be especially useful in situations where a diagnosis remains elusive after standard testing or in situations in which the cause of a disease outbreak is unknown."
Details were published in the September 22, 2015, online edition of the journal Genome Research.
Related Links:
Washington University School of Medicine
454 Roche NimbleGen