Raman Imaging Probes for Detecting Enzyme Activities Could Aid Disease Diagnosis
Posted on 02 Jun 2023
Enzymes play a crucial role in various biological processes, which makes them suitable biomarkers for disease detection. For instance, cancer diagnosis often uses fluorescence imaging to identify cancer-related enzymes that have multiplied in cells affected by the disease. Given the heterogeneous nature of tumor tissues, being able to detect multiple enzyme activities at once could enhance the precision of cancer visualization and diagnosis. However, detecting multiple enzyme activities using fluorescence imaging can be challenging in heterogeneous tumor tissues and other complex biological phenomena. Raman spectral imaging, with its narrower spectral width, provides a promising alternative for multiplex biological imaging with molecular probes. Over time, a number of functional and activatable Raman probes (dyes) have been developed for bioanalyte detection, but their use for enzyme activity detection has been limited. In addition, prior design strategies have been unsuccessful in controlling the diffusion of enzyme-generated hydrolysis product of these probes, making it difficult to identify areas with targeted enzyme activity in tissues.
Now, a team of researchers led by Tokyo Institute of Technology (Tokyo Tech, Tokyo, Japan) has come up with a novel molecular design approach, taking cues from aggregation-based fluorescent probes. The team has developed activatable Raman probes based on 9CN-rhodol using a novel mechanism for Raman signal activation. The researchers have shown that Raman imaging has more potential for detecting multiple enzyme activities compared to fluorescence imaging. This innovative strategy allows the creation of highly activatable Raman probes that possess strong aggregation and multiplexing ability, providing a promising tool for expanding the range of Raman probes that can detect multiple enzyme activities in heterogeneous biological tissues.
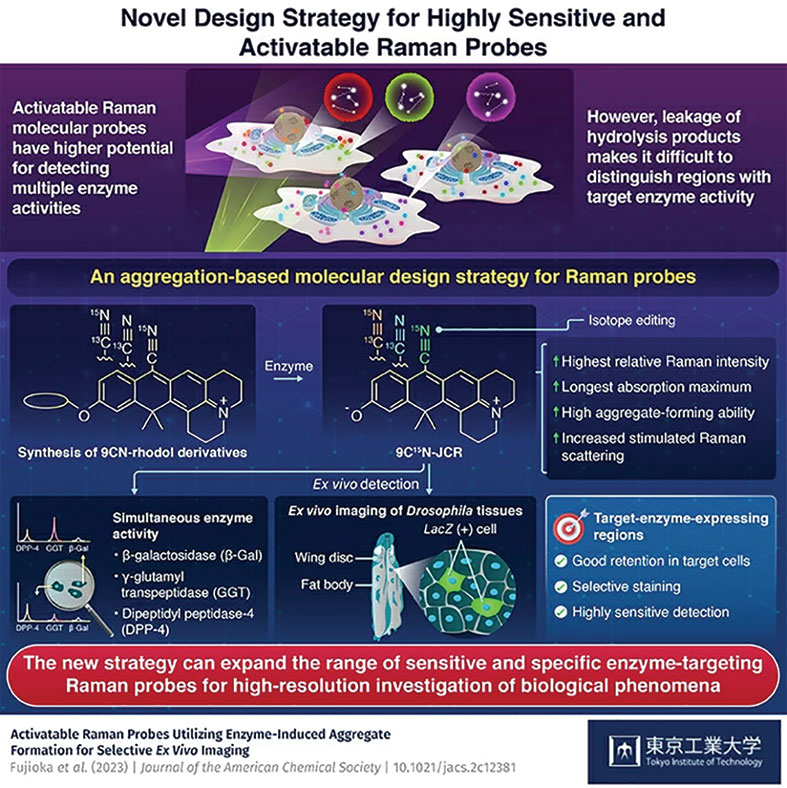
The researchers first synthesized 9CN-rhodol derivatives and chose two, 9CN-JR and 9CN-JCR, to design the activatable Raman probes. They tested the enzyme detection capabilities of both probes in live cells, using a dual-color stimulated Raman scattering (SRS) imaging technique. Among the two, 9CN-JCR outperformed and proved to be the more effective probe for multiplexing. The team then isotope-labeled the nitril group of 9CN-JCR scaffold with Carbon-13 (13C) and Nitrogen-15 (15N), and developed two new isotope edited 9CN-JCR probes for γ-glutamyl transpeptidase and dipeptidyl peptidase-4 enzymes. These 9CN-JCR-based probes successfully detected all enzyme activities simultaneously in the live cell culture.
Additionally, the probes enabled ex vivo imaging of specific cell areas showing targeted enzyme activity in Drosophila wing disk and fat body. The high spatial selectivity and sensitivity exhibited by the 9CN-JCR probes were attributed to the electronic pre-resonance effect of the scaffold dye and aggregate formation of the hydrolysis products formed by probe-cell interaction. These rhodol-based probes could aggregate when reacting with enzymes, which enhanced their intracellular retention and boosted the SRS signal intensity during enzyme detection. Overall, the approach presented in this study could support the development of highly specific activatable Raman probes for simultaneous detection of multiple enzyme activities.
"Our aggregation-based molecular design strategy for Raman probes will offer substantial advantages for applications involving the investigation of enzyme activity associated with diseases and essential biological activities," said Professor Mako Kamiya of Tokyo Tech who led the research.
Related Links:
Tokyo Tech