Natural Killer Cell-Associated Markers Investigated in Gastric Cancer
By LabMedica International staff writers
Posted on 11 Jan 2022
Gastric cancer (GC) is one of the leading causes of cancer-related deaths worldwide. Although the survival rates of GC patients have increased with proper screening systems, standardized surgical protocols, and development of chemotherapy regimens, the general outcome of GC patients, especially those in advanced stages, remains poor.Posted on 11 Jan 2022
The importance of the tumor immune microenvironment (TME) has been well-established in the last two decades. Various immune cell populations and stromal cells in the TME and tumor cells play an important role by interacting with each other, leading to tumor suppression or progression. Characterizing the interactions between immune cells and tumor cells is critical to understand the TME.
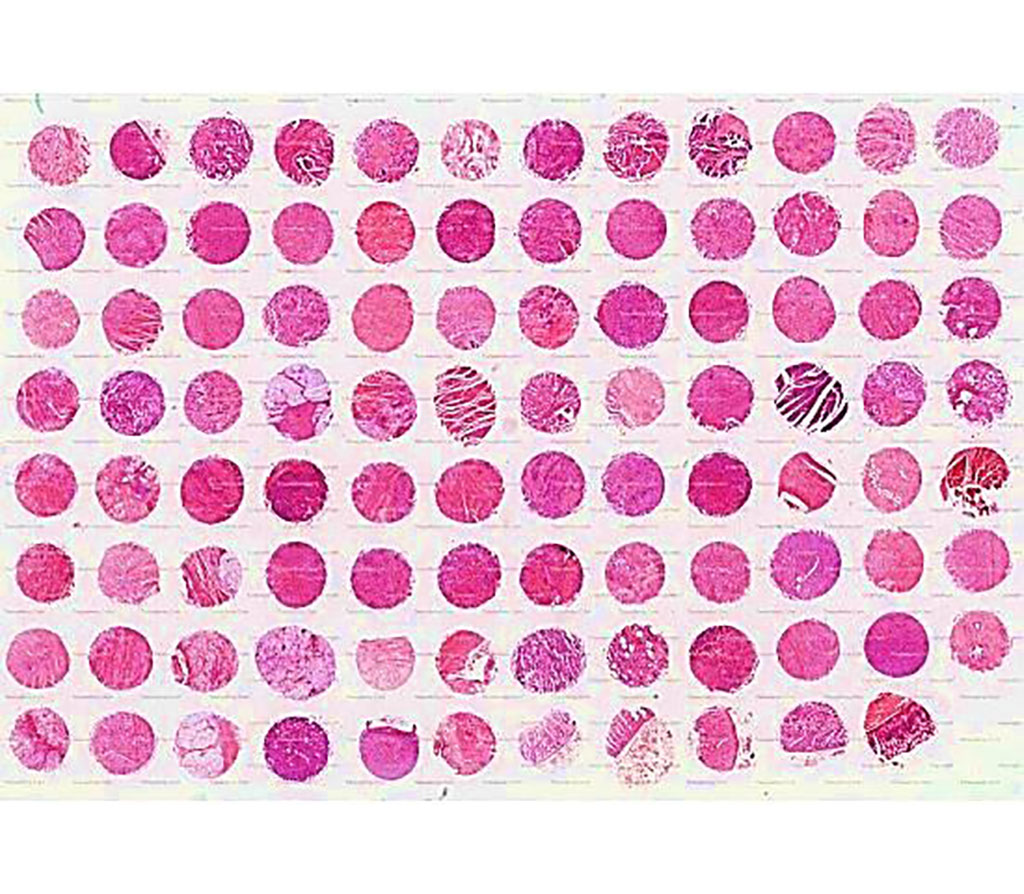
Image: Advanced stage of stomach cancer with stomach tissue array, including pathology grade, malignant tumors (TNM) and clinical stage (Photo courtesy of SuperBioChips Laboratories)
Clinical Scientists at the Seoul National University College of Medicine (Seoul, Republic of Korea) recruited for a study a total of 55 cases of consecutive stage II-III GC surgically resected between 2006 and 2008. The formalin-fixed, paraffin-embedded (FFPE) tissue samples were reviewed, and representative tissue areas were dissected for the construct a tissue microarray (TMA). The team performed multiplex immunohistochemistry (mIHC) on the tissue microarrays slides. A total of 11 antibodies including CD57, NKG2A, CD16, HLA-E, CD3, CD20, CD45, CD68, CK, SMA, and ki-67 were used. CD45 + CD3-CD57 + cells were considered as CD57 + NK cells.
For molecular classification of the GC samples, conventional immunohistochemistry (IHC) for E-cadherin and p53 was performed on 3-μm-thick TMA slides using an automated BenchMark XT immunostainer (Ventana Medical Systems, Tucson, AZ USA). All implementations and analysis of mIHC were performed on the SuperBioChips (SuperBioChips Laboratories, Seoul, Korea). Polymerase chain reaction (PCR) amplification of the extracted DNA from tumor and normal cells was performed and the PCR products were analyzed using a DNA autosequencer, the ABI 3731 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
The investigators reported that among CD45 + immune cells, the proportion of CD57 + NK cell was the lowest (3.8%), whereas that of CD57 + and CD57- T cells (65.5%) was the highest, followed by macrophages (25.4%), and B cells (5.3%). CD57 + NK cells constituted 20% of CD45 + CD57 + immune cells while the remaining 80% were CD57 + T cells. The expression of HLA-E in tumor cells correlated with that in tumoral T cells, B cells, and macrophages, but not CD57 + NK cells. The higher density of tumoral CD57 + NK cells and tumoral CD57 + NKG2A + NK cells was associated with inferior survival.
The authors concluded that although the number of CD57 + NK cells was lower than that of other immune cells, CD57 + NK cells and CD57 + NKG2A + NK cells were significantly associated with poor outcomes, suggesting that NK cell subsets play a critical role in GC progression. NK cells and their inhibitory receptor, NKG2A, may be potential targets in GC. The study was published on December 24, 2021 in the Journal of Translational Medicine.
Related Links:
Seoul National University College of Medicine
Ventana Medical Systems
SuperBioChips Laboratories
Applied Biosystems