Extrachromosomal Circular DNA Drives Oncogenic Genome Remodeling in Neuroblastoma
By LabMedica International staff writers
Posted on 02 Jan 2020
Circularized DNA falling outside of linear chromosomes may serve as a recurrent source of somatic rearrangements in neuroblastoma, a pediatric cancer affecting immature cells in the sympathetic nervous system.Posted on 02 Jan 2020
While past studies have pointed to a role for circularized, extrachromosomal MYCN oncogene sequences in neuroblastoma, the full suite and the frequency of somatic mutations involving small or large stretches of circularized extrachromosomal DNA amplifications had not been fully explored.
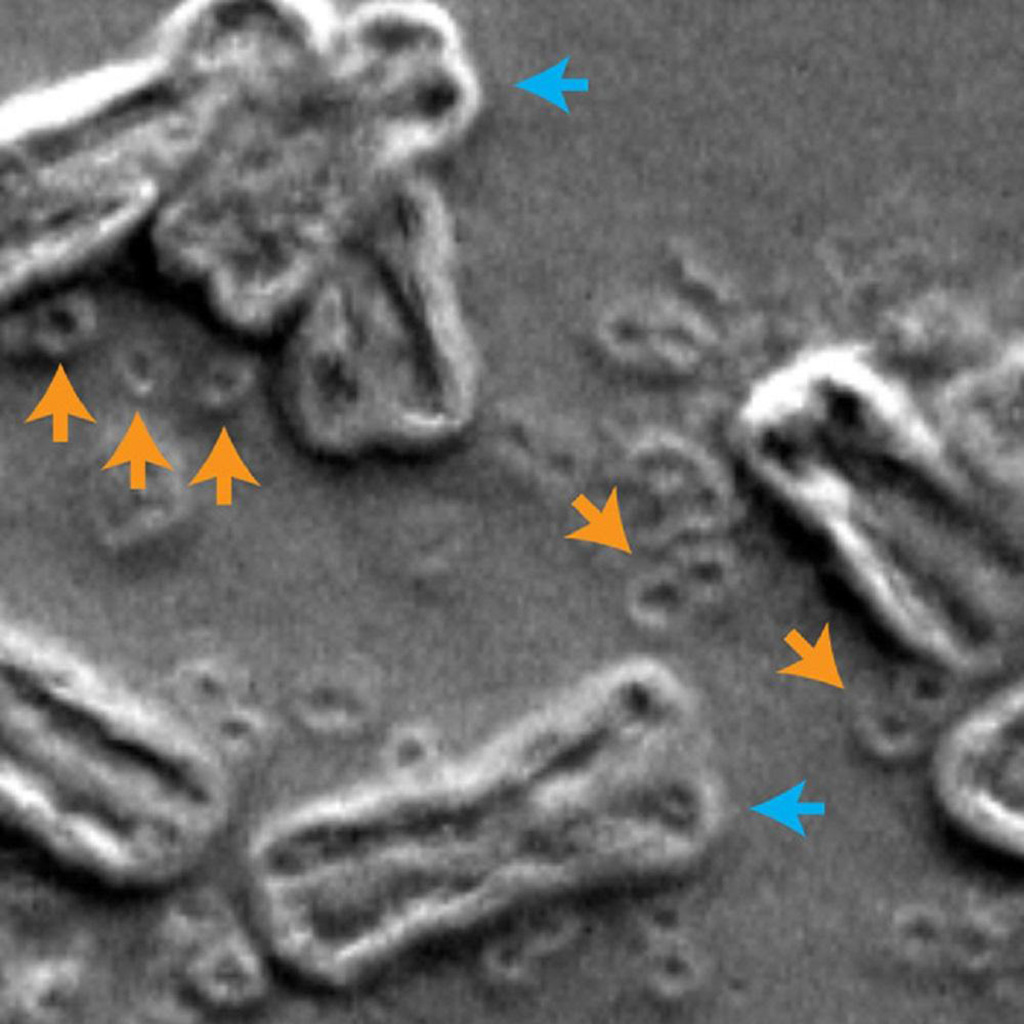
Image: Scanning electron micrograph of inside the nucleus of a cancer cell, chromosomes are indicated by blue arrows and circular extrachromosomal DNA are indicated by orange arrows (Photo courtesy of Paul S. Mischel, MD, UC San Diego)
An international team of scientists collaborating with those at Charité-Universitätsmedizin Berlin (Berlin, Germany) profiled matched tumor and normal blood samples from 93 neuroblastoma patients using whole-genome sequencing and an algorithm that uncovers circularized DNA based on paired read orientation, uncovering preliminary evidence for complex and relatively frequent extrachromosomal DNAs (ecDNAs) in neuroblastoma.
The team to take a closer look at these sequences using a modified version of circle sequencing (Circle-seq) in 21 of the neuroblastoma tumors, making it possible to enrich for circularized DNA. The circularized sequences were mapped back to their original sites in the genome using additional long-read and single-molecule real-time sequences, the investigators explained, and they validated candidate DNA circles with polymerase chain reaction (PCR) and Sanger sequencing.
Together, these approaches uncovered almost 5,700 small extrachromosomal circular DNAs per tumor, on average, and an average of 0.82 large, copy number-amplified extrachromosomal circular DNA sequences. Even so, the team's follow-up analyses, including RNA sequencing experiments, indicated that rearrangements stemming from extrachromosomal circular DNA from MYCN and other genes may be a recurrent and ongoing source of new mutations through a multi-hit model in neuroblastoma.
The authors concluded that they had demonstrated that the majority of genomic rearrangements in neuroblastoma involve circular DNA, challenging the current understanding about cancer genome remodeling. They envision that their findings extend to other cancers and that further detailed analyses of circle-derived rearrangements will shed new insights into our understanding of cancer genome remodeling. The study was published on December 16, 2019 in the journal Nature Genetics.
Related Links:
Charité-Universitätsmedizin Berlin