Protein Linked to Aggressive Skin Cancer
By LabMedica International staff writers
Posted on 10 Jul 2019
Almost 300,000 people worldwide develop malignant melanoma each year. The disease is the most serious form of skin cancer and the number of cases reported annually is increasing, making skin cancer one of Sweden's most common forms of cancer.Posted on 10 Jul 2019
Over the past ten years, new treatment alternatives that use different methods to strengthen the immune system or attack specific cancer cells have been developed for patients with metastatic skin cancer. The introduction of these treatments is due to an increased understanding of how melanoma develops, but there is still a lack of knowledge about how the tumor cells spread to other parts of the body.
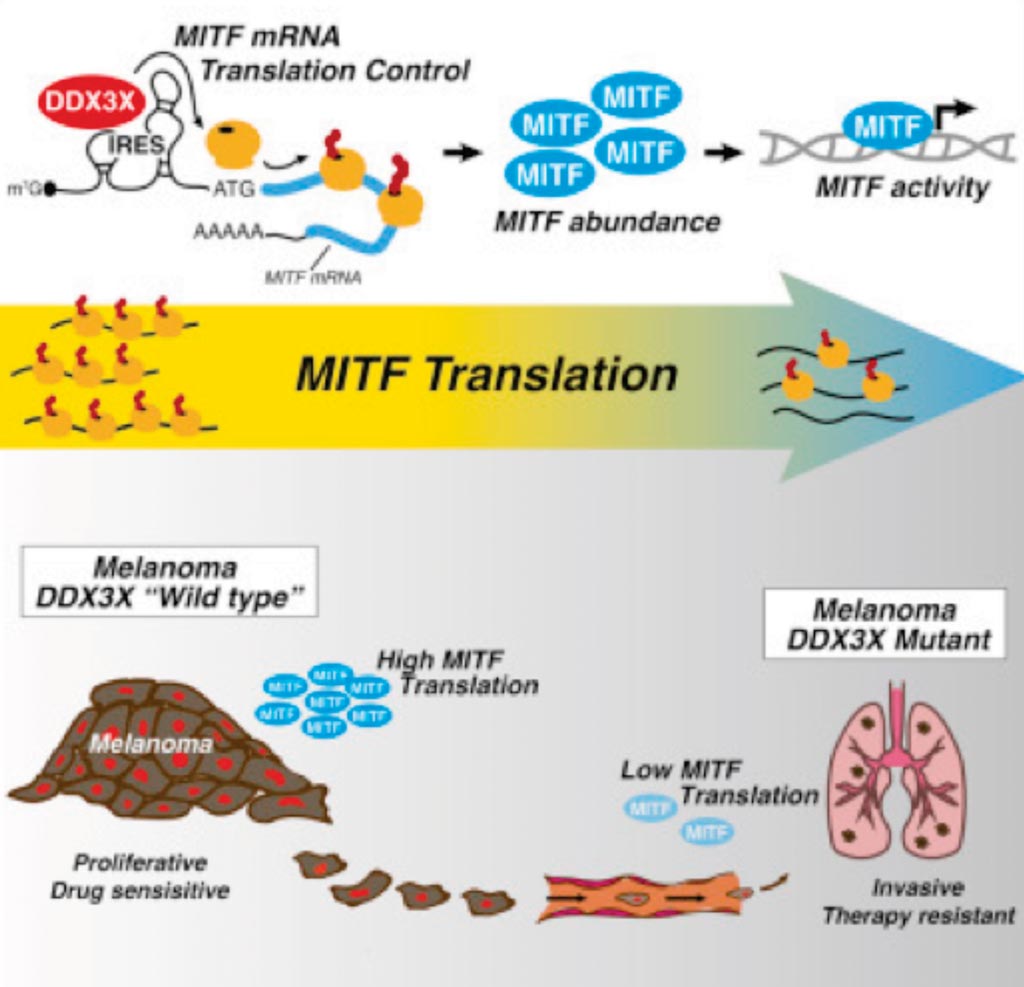
Image: A diagram of how the X-Linked DDX3X RNA helicase dictates translation reprogramming and metastasis in melanoma (Photo courtesy of Lund University).
A team of scientists mainly from Lund University (Lund, Sweden) collected lymph node metastatic tissue from 124 patients (48 females, 73 males and three missing information) with a regional metastatic disease and average age at diagnosis of 61 (range 25-86) was used in the quantitative polymerase chain reaction (qPCR) analysis. The team used a multiplicity of methods to obtain their results.
The investigators uncovered an ATP-Dependent RNA Helicase (DDX3X)-driven post-transcriptional program that dictates melanoma phenotype and poor disease prognosis. Through an unbiased analysis of translating ribosomes, they identified the microphthalmia-associated transcription factor, MITF, as a key DDX3X translational target that directs a proliferative-to-metastatic phenotypic switch in melanoma cells. Mechanistically, DDX3X controls MITF mRNA translation via an internal ribosome entry site (IRES) embedded within the 5′ UTR. Through this exquisite translation-based regulatory mechanism, DDX3X steers MITF protein levels dictating melanoma metastatic potential in vivo and response to targeted therapy.
The team noted that that the DDX3X protein does not affect whether or not one develops malignant melanoma, but that it plays a considerable role in the aggressiveness of the tumor. The patient's level of DDX3X can therefore serve as a biomarker for predicting how intractable the disease will be. Göran B. Jönsson, MD, a professor of oncology and pathology, and a senior author of the study, said, “The activity of the MITF gene determines the melanoma cells' specific characteristics, which are then linked to the disease prognosis. The lower the level of DDX3X protein the patient has in the tumor cell, the more aggressive the disease and the worse the prognosis will be.” The study was published on June 18, 2019, in the journal Cell Reports.
Related Links:
Lund University