Standard Clinical Assays Obscure Breast Cancer Subtype Diversity
By LabMedica International staff writers
Posted on 11 May 2015
The classification systems that categorize breast cancers based on estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) expression levels may obscure the heterogeneity of other key tumor features.Posted on 11 May 2015
Some breast tumors that share a single clinical label, such as triple-negative breast cancer (TNBC), may in fact represent a diverse collection of molecular subtypes and many breast cancer treatment decisions hinge on whether tumors test positive or negative for ER, PR, and/or HER2, yet the criteria for interpreting a test result can vary.
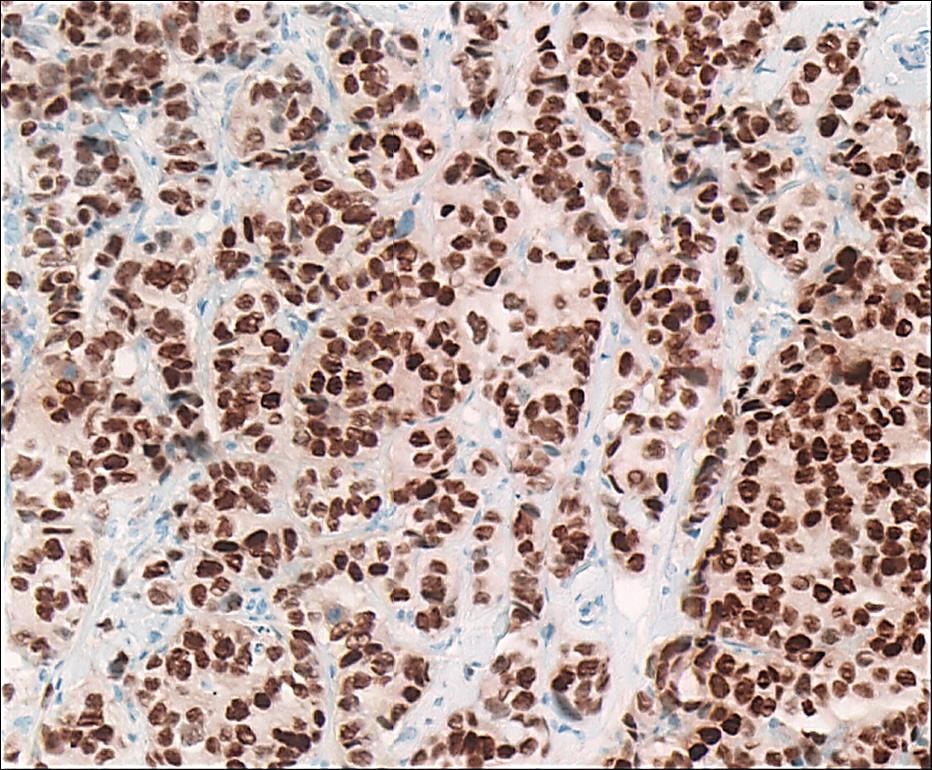
Image: Estrogen receptor (ER)-positive breast cancer surgical pathology specimen stained for ER by immunohistochemistry. Most of the cancer cell nuclei stain dark brown, strongly positive for ER (Photo courtesy of Ronald S. Weinstein, MD).
Scientists at the University of North Carolina (Chapel Hill, NC, USA) and a large team of international collaborators studied 1,557 breast tumor samples that were tested for ER, PR, and HER2 expression by standard clinical assays. The same tumors were also tested for their molecular features and classified into one of five molecular subtypes: Luminal A, Luminal B, HER2-Enriched, Basal-like, and Normal-like.
These breast tumors were centrally reviewed in three different trials for quantitative ER, PR, and HER2 expression by immunohistochemistry (IHC) stain and by reverse transcription-quantitative polymerase chain reaction (RT-qPCR), with intrinsic subtyping by the gene set PAM50 RT-qPCR assay. HER2 expression was determined by IHC, and the amplification ratio was determined by fluorescent in situ hybridization (FISH) and also by chromogenic in situ hybridization (CISH).
Among TNBCs with less than 1% hormone receptor (HR) staining, the most common molecular subtype was Basal-like (73%), followed by HER2-Enriched (17%). By comparison, TNBC tumors with borderline HR staining had a much wider mix of molecular subtypes, including Luminal A/B (44%), HER2-Enriched (31%), and Basal-like (18%). Among the 228 basal-like tumors, 93.4% (213 of 228) had less than 1% ER or PR staining by IHC.
Lisa A. Carey, MD, the senior author of the study, said, “Including tumors with borderline HR staining in the definition of triple-negative breast cancer significantly diminished the proportion of Basal-like molecular subtypes. The optimal threshold for enriching for Basal-like breast cancer is less than 1% for either hormone receptor. Our findings show that borderline HR-expressing tumors are heterogeneous and do not fit well into distinct molecular categories. This raises the question of whether ‘borderline’ HR staining should instead be considered ‘indeterminate’ and requires additional assays to clarify the underlying biology.” The study was published on April 23, 2015, in the journal the Oncologist.
Related Links:
University of North Carolina










.jpg)