Histology Expression Predictor Assay Eliminates Subjectivity from Lung Cancer Diagnostics
By LabMedica International staff writers
Posted on 29 Jul 2013
A gene expression-based predictor assay for lung cancer FFPE (formalin-fixed, paraffin-embedded) specimens was able to identify a variety of cancers with accuracy and precision similar to that of microscopic examination by pathologists. Posted on 29 Jul 2013
FFPE tissues are the most widely available specimens for retrospective clinical studies of disease mechanisms. These archived materials provide a valuable source of stable nucleic acids for gene expression analysis, using real-time quantitative reverse transcription-PCR (qRT-PCR) or microarray analysis. As the use of PCR technology has become more prevalent in molecular testing, it has enhanced the clinical utility of FFPE tissues. However, the recovery of quality RNA from FFPE specimens is often problematic, as the fixation process causes cross-linkage between nucleic acids and proteins, and covalently modifies RNA by the addition of monomethyl groups to the bases. As a result, the molecules are rigid and susceptible to mechanical shearing, and the cross-links may compromise the use of RNA as a substrate for reverse transcription. Therefore, in order to utilize FFPE tissues as a source for gene expression analysis, a reliable method is required for extraction of RNA from the cross-linked matrix.
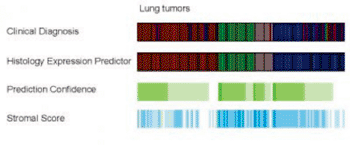
Image: Histological types are labeled by different colors in the clinical diagnosis and histology expression-predictor tracks. Prediction confidence and stromal score are shaded green to white to blue indicating high to low values. The highest quality predictions exhibit high confidence and low stromal scores (Photo courtesy of the University of North Carolina).
Lung cancer histologic diagnosis is clinically relevant because there are treatment indications and contraindications that are histology-specific. In practice, histologic diagnosis can be challenging depending on the success of sampling and on various tumor characteristics. In addition, the subjective nature of microscopic examination can lead to disagreements among pathologists reviewing the same specimens. Investigators at the University of North Carolina (Chapel Hill, USA) and the University of Utah (Salt Lake City, USA) hope to have solved some of these problems by developing a gene expression-based predictor of lung cancer histology for FFPE specimens.
Genes predictive of lung cancer histologies were derived from published cohorts that had been profiled by microarrays. Expression of these genes was measured by RT-qPCR in FFPE samples from a cohort of 442 lung cancer patients. A histology expression predictor (HEP) was developed using RT-qPCR expression data for adenocarcinoma, carcinoid, small cell carcinoma, and squamous cell carcinoma.
In cross-validation, the HEP exhibited mean accuracy of 84%. The HEP was compared with pathologist diagnoses on the same tumor block specimens, and the HEP yielded accuracy and precision similar to the pathologists.
"Our predictor identifies the major histologic types of lung cancer in paraffin-embedded tissue specimens, which is immediately useful in confirming the histologic diagnosis in difficult tissue biopsy specimens," said contributing author Dr. Neil Hayes, associate professor of medicine at the University of North Carolina. "As we learn more about the genetics of lung cancer, we can use that understanding to tailor therapies to the individual's tumor. Gene expression profiling has great potential for improving the accuracy of the histologic diagnosis. Historically, gene expression analysis has required fresh tumor tissue that is usually not possible in routine clinical care. We desperately needed to extend the analysis of genes (aka RNA) to paraffin samples that are routinely generated in clinical care, rather than fresh frozen tissue. That is the major accomplishment of the current study and one of the first large scale endeavors in lung cancer to show this is possible."
A detailed description of the histology expression-predictor assay was published in the July 2013 issue of the Journal of Molecular Diagnostics.
Related Links:
University of North Carolina
University of Utah