Ultrasensitive Method Detects Low-Frequency Cancer Mutations
Posted on 18 Dec 2025
Liquid biopsy has emerged as a promising approach for cancer detection and treatment monitoring, although its clinical impact has been limited by the extremely low levels of tumor-derived DNA circulating in blood. These rare fragments are often obscured by abundant normal DNA and background sequencing errors, making early cancer detection and monitoring of residual disease particularly challenging. A new approach now demonstrates that selectively removing normal DNA before sequencing can dramatically amplify rare cancer mutations, enabling reliable detection even at frequencies previously considered undetectable.
Researchers from Korea University College of Medicine (Seoul, South Korea), in collaboration with multiple research partners, have developed a CRISPR-based sequencing enrichment strategy designed to improve the sensitivity of liquid biopsy testing while reducing background noise and sequencing costs. The method relies on an engineered high-fidelity CRISPR enzyme capable of distinguishing single-base differences in DNA.
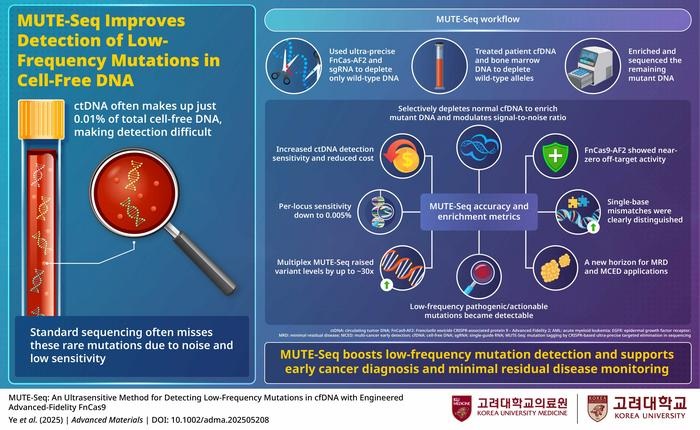
By selectively cleaving perfectly matched normal DNA and sparing mutant sequences, the approach enriches circulating tumor DNA before sequencing, allowing rare variants to become detectable above intrinsic error rates. This selective enrichment step functions as a built-in noise-reduction process that can be integrated into standard sequencing workflows, with or without unique molecular identifiers.
The strategy is compatible with multiplex analysis, enabling simultaneous interrogation of multiple cancer-related mutation hotspots within a single assay. Because the enrichment occurs before sequencing, the method improves efficiency and reduces the need for excessive sequencing depth, making it more practical for routine clinical use.
The technique was validated using Sanger sequencing, next-generation sequencing, and cell-free DNA reference materials. Across these tests, variant allele frequencies increased by tens of times, enabling the detection of mutations present at approximately 0.005%. In patients with acute myeloid leukemia, weak NRAS mutation signals associated with minimal residual disease became clearly detectable.
Multiplex testing across EGFR and KRAS hotspots improved concordance between plasma and tumor tissue in patients with non-small cell lung cancer and pancreatic cancer, including early-stage cases. Overall sensitivity improvements ranged from twenty to sixtyfold while maintaining high specificity, with a calculated detection limit of 0.034% variant allele frequency using 50 ng of input DNA. These results suggest the approach can reliably identify minute cancer-associated mutations that are typically lost in sequencing noise.
The findings, published in Advanced Materials, support broad applications in multi-cancer early detection, minimal residual disease monitoring, and tracking treatment-emergent resistance mutations. Future work will focus on translating this strategy into clinically deployable diagnostic panels and expanding validation across larger patient cohorts.
“Our findings suggest that this method has considerable potential for developing diagnostic panels aimed at detecting multiple low-frequency circulating tumor DNA mutations for early cancer detection, companion diagnostics, and minimal residual disease monitoring,” said Professor Junseok W Hur, lead investigator of the study.
Related Links:
Korea University College of Medicine