Single Metagenomic Next-Generation Sequencing Test Can Detect All Infectious Pathogens
Posted on 12 Nov 2024
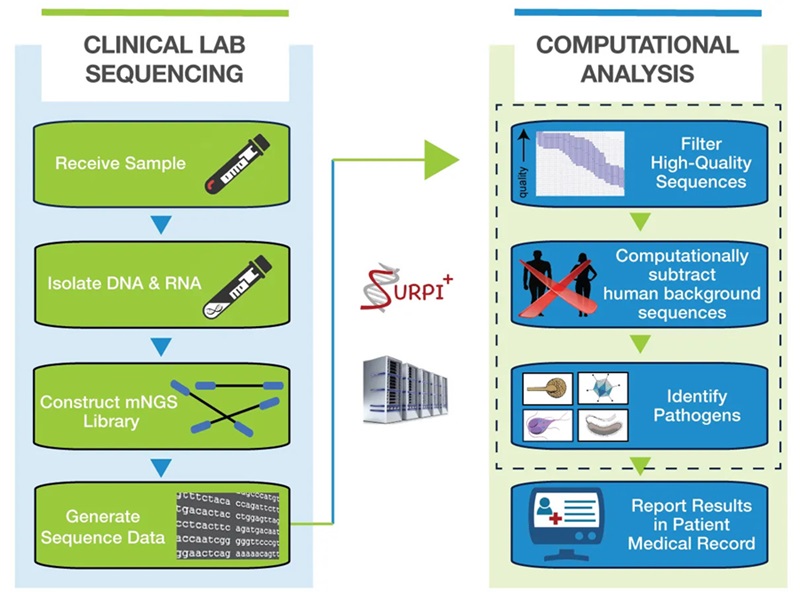
Metagenomic next-generation sequencing (mNGS) is a comprehensive sequencing method that analyzes all nucleic acids (DNA and RNA) in a clinical sample at high depth, generating 10-20 million sequences per sample. This approach can be applied to any type of clinical sample, including cerebrospinal fluid, plasma, respiratory secretions, urine, stool, or tissue. Now, mNGS offers the potential to diagnose a wide range of infectious agents—viruses, bacteria, fungi, and parasites—through a single test.
Researchers at the UCSF Center for Next-Gen Precision Diagnostics (San Francisco, CA, USA) have developed an mNGS test capable of detecting sequence reads for all pathogens, helping to identify the cause of an infection. Initially, the UCSF team developed an mNGS test to identify pathogens in cerebrospinal fluid for meningitis and encephalitis diagnosis. This was followed by the development of a clinical plasma mNGS test designed to detect DNA viruses, bacteria, fungi, and parasites responsible for sepsis and disseminated infections in other organs. To analyze mNGS data quickly for pathogen identification, the team employs SURPI+ (SURPI PLUS), a clinical bioinformatics software pipeline.
In research published in Nature Medicine, the team demonstrated that their mNGS test for cerebrospinal fluid accurately identified 86% of neurological infections. They are now working on validating clinical tests for detecting all microorganisms (bacteria, viruses, fungi, and parasites) causing pneumonia in respiratory fluids like bronchoalveolar lavage or tracheal aspirates. Additionally, in a related study published in Nature Communications, the team used mNGS to identify pneumonia-causing pathogens in respiratory fluids and automated the process to generate faster results.













