Noninvasive Blood Test Predicts Rejection After Heart Transplant
Posted on 06 Jun 2022
Heart transplantation (HTx) is the definitive treatment option for patients with advanced heart failure. Despite continued advances in post-transplant outcomes, allograft rejection and allograft injury remain impediments to post-transplant survival.
Endomyocardial biopsy and histopathology remain the principal surveillance tools for rejection after heart transplant since they were developed decades ago. However, endomyocardial biopsy is invasive and expensive, and histopathology reads are prone to inter-observer variability.
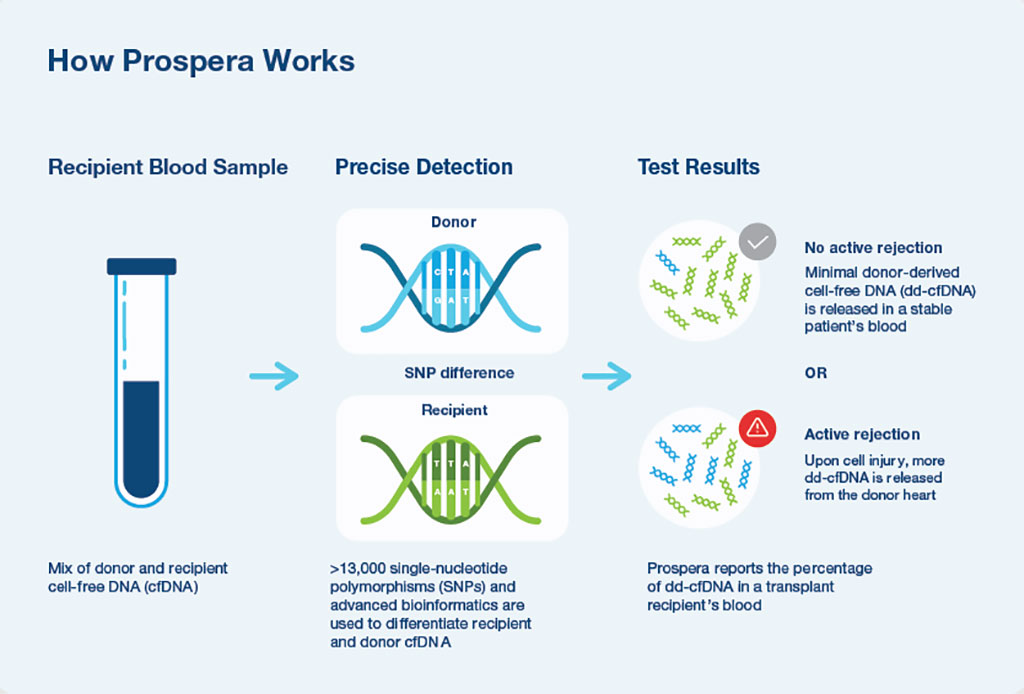
Physicians at UC San Diego Health (San Diego, CA, USA) and their colleagues examined the performance characteristics of a novel test for detecting acute rejection (AR) in adult HTx recipients. Plasma samples with contemporaneous endomyocardial biopsies (EMBs) were obtained from HTx recipients. A total of 811 samples from 223 patients with dd-cfDNA testing and contemporaneous EMB were eligible for the study.
A clinically available SNP-based massively multiplexed-PCR dd-cfDNA assay was used to measure dd-cfDNA fraction. dd-cfDNA fractions were compared with EMB-defined rejection status and test performance was assessed by constructing ROC curves and calculating accuracy measures. Laboratory testing involved cfDNA extraction and library preparation using the Prospera test (Natera Inc., Austin, TX, USA). This was followed by cfDNA amplification using massively multiplexed-PCR, targeting over 13,000 single nucleotide polymorphisms designed to maximize the number of informative SNPs across ethnicities and next-generation sequencing of the resultant amplicons, with sequencing performed on the NextSeq500 (Illumina, San Diego, USA) on rapid run with an average of 14 to 15 million reads per sample.
The scientists reported that AR was observed in 49 biopsy matched samples from 35 patients while 762 samples from 210 patients did not show AR. Median dd-cfDNA fraction was significantly higher in samples with a matched biopsy showing AR (median 0.58%, IQR, 0.13%-1.68%) compared to samples where matched biopsies did not show AR. ROC analysis produced an area under the curve (AUC-ROC) of 0.86. Defining samples with dd-cfDNA fraction ≥0.15% as AR yielded 78.5% sensitivity and 76.9% specificity. Positive and negative predictive values were 25.1% and 97.3% respectively, calculated using the cohort AR prevalence of 9.0% with adjustment for repeat samples.
The authors concluded that their study affirms an association between elevated levels of dd-cfDNA and histologic evidence of rejection after heart transplant, and extends previous findings showing that dd-cfDNA is a valuable biomarker of allograft health. The study was published originally published on April 9, 2022 in the Journal of Heart and Lung Transplantation.
Related Links:
UC San Diego Health
Natera Inc
Illumina