'Fingerprint' Machine Learning Technique Identifies Different Bacteria in Seconds
Posted on 08 Mar 2022
A synergistic combination of surface-enhanced Raman spectroscopy and deep learning serves as an effective platform for separation-free detection of bacteria in arbitrary media.
Researchers at the Korea Advanced Institute of Science and Technology (KAIST, Daejeon, Korea) have demonstrated a quicker, more accurate process for bacterial identification which can take hours and often longer, despite time being precious when diagnosing infections and selecting appropriate treatments. By teaching a deep learning algorithm to identify the “fingerprint” spectra of the molecular components of various bacteria, the researchers could classify various bacteria in different media with accuracies of up to 98%.
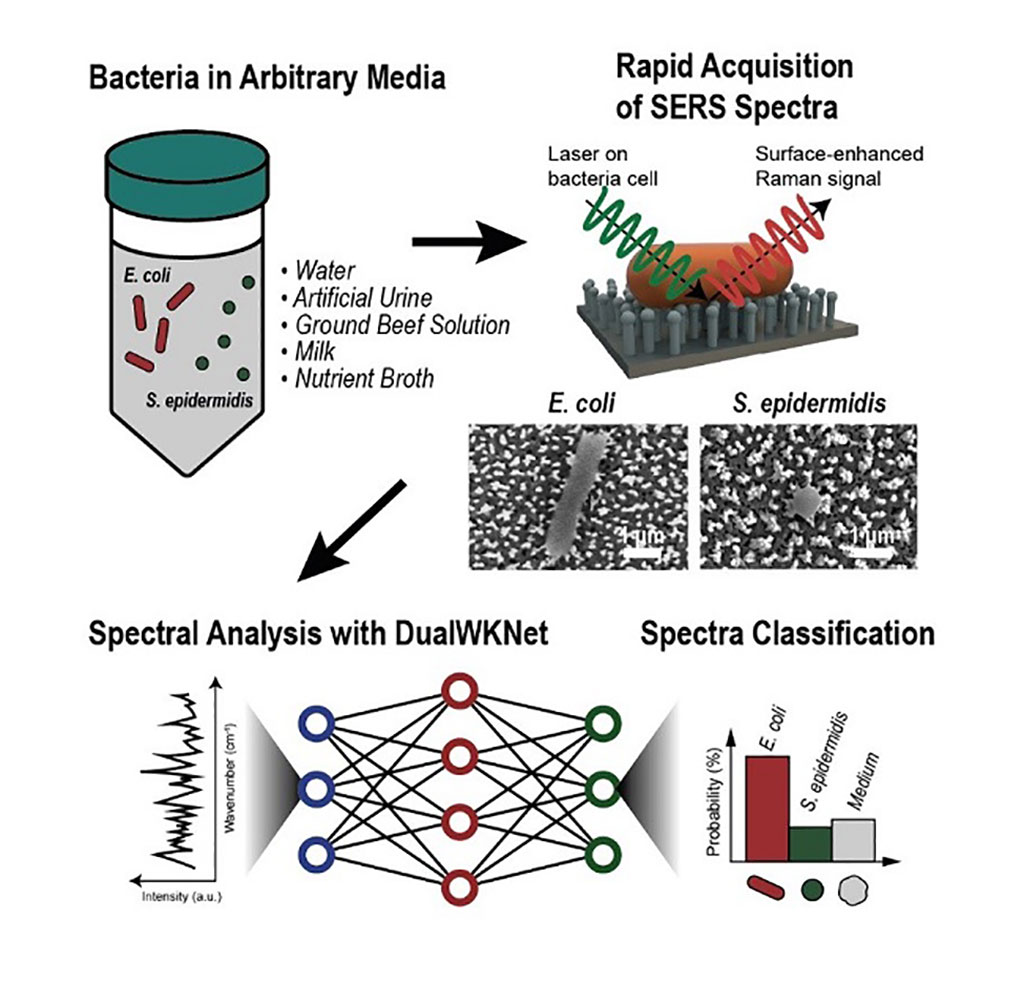
Bacteria-induced illnesses, those caused by direct bacterial infection or by exposure to bacterial toxins, can induce painful symptoms and even lead to death, so the rapid detection of bacteria is crucial to prevent the intake of contaminated foods and to diagnose infections from clinical samples, such as urine. Raman spectroscopy sends light through a sample to see how it scatters. The results reveal structural information about the sample - the spectral fingerprint - allowing researchers to identify its molecules. Surface-enhanced Raman spectroscopy (SERS) places sample cells on noble metal nanostructures that help amplify the sample’s signals.
However, it is challenging to obtain consistent and clear spectra of bacteria due to numerous overlapping peak sources, such as proteins in cell walls. To parse through the noisy signals, the researchers have implemented an artificial intelligence method called deep learning that can hierarchically extract certain features of the spectral information to classify data. They specifically designed their model, named the dual-branch wide-kernel network (DualWKNet), to efficiently learn the correlation between spectral features. Such ability is critical for analyzing one-dimensional spectral data, according to the researchers. The researchers now plan to use their platform to study more bacteria and media types, using the information to build a training data library of various bacterial types in additional media to reduce the collection and detection times for new samples.
“We developed a meaningful universal platform for rapid bacterial detection with the collaboration between SERS and deep learning,” said Professor Sungho Jo from the School of Computing. “We hope to extend the use of our deep learning-based SERS analysis platform to detect numerous types of bacteria in additional media that are important for food or clinical analysis, such as blood.”
Related Links:
KAIST













