Targeted RNA-Sequencing Panel Detects Gene Fusions in Solid Tumors
By LabMedica International staff writers
Posted on 07 Dec 2021
Increasingly, rearrangements resulting in gene fusions have been revealed in multiple tumor types across diverse organ systems. These gene fusions can drive tumorigenesis by altering gene expression and activity. Timely and reliable detection of fusions provides useful information to the pathologist and clinical team.Posted on 07 Dec 2021
Molecular techniques, such as fluorescence in situ hybridization (FISH), reverse transcription PCR (RT-PCR), and Sanger and next-generation sequencing (NGS), have enabled discovery of characteristic fusions associated with particular tumors. With FISH and RT-PCR, the genes involved must be known or suspected, and usually only a few prioritized assays are run, due to cost and time limitations. These methods are not suitable for the identification of novel fusion partners.
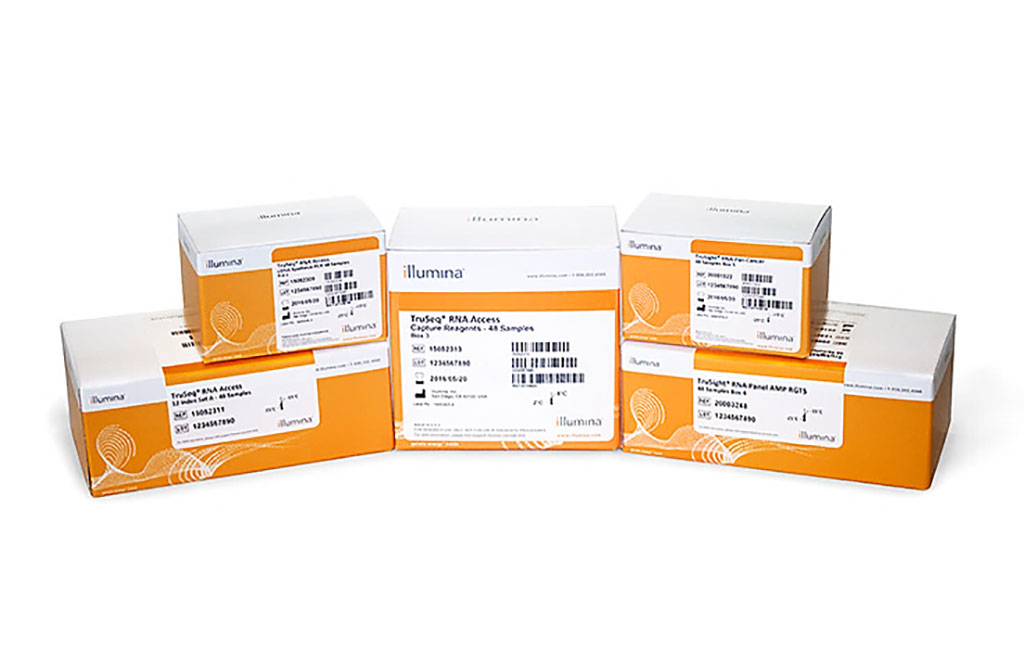
Image: The TruSight RNA Fusion Panel kit provides comprehensive gene fusion detection in formalin-fixed, paraffin-embedded (FFPE) and other cancer samples (Photo courtesy of Illumina)
Clinical Scientists at the Washington University School of Medicine (St. Louis, MO, USA) included 153 sarcoma and carcinoma cases submitted for FISH, RT-PCR, or DNA-based NGS testing during diagnostic workup between 2013 and 2019 that were identified retrospectively from their institutional database. Hematoxylin and eosin–stained slides and paraffin blocks were screened for adequate tissue (at least enough for a 1-mm punch, with ≥50% tumor cellularity in the punched area).
Deparaffinization and RNA extraction were performed using RecoverALL Total Nucleic Acid Isolation Kit for FFPE (Thermo Fisher Scientific, Waltham, MA). Quality of RNA and input amount were determined by Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), with DV200 values, per TruSight RNA Fusion Panel kit protocol (Illumina, San Diego, CA, USA). Library preparation was performed using the TruSight RNA Fusion Panel kit. Sequencing was performed on the Illumina MiSeq and NextSeq 550 systems, using paired-end (2 × 75 nucleotides) reads.
The team reported that a total of 138 cases were successfully sequenced with adequate quality control metrics (90%). There were 110 sarcomas, with 31 sarcomas not otherwise specified, 25 Ewing sarcomas, and 13 synovial sarcomas, among others. A total of 101 of 138 (73%) cases were concordant by RNA sequencing and the prior test method. RNA sequencing identified an additional 30 cases (22%) with fusions that were not detected by conventional methods. In seven cases (5%), the additional fusion information provided by RNA sequencing would have altered diagnosis and management. A total of 19 (14%) novel fusion pairs, not previously described in the literature, were discovered.
The authors concluded that fusion analysis is a beneficial ancillary test that provides significant clinical value to the pathologist and treatment team. Targeted RNA sequencing on FFPE tissue is one method that enables the sensitive detection of multiple potential fusion genes simultaneously. This test could be performed at the time of pathologic examination in biopsy or resection specimens to aid in accurate diagnosis and clinical management. The study was published on December 1, 2021 in the Journal of Molecular Diagnostics.
Related Links:
Washington University School of Medicine
Thermo Fisher Scientific
Agilent Technologies
Illumina