Novel Tool Developed to Detect and Identify Pathogens
By LabMedica International staff writers
Posted on 28 Sep 2021
Pathogens in clinical or wildlife settings samples of blood or saliva, for example, are particularly challenging to isolate, since they can easily make up less than one one-millionth of a sample, especially in early stages of an infection, when concentrations are still low and detection is most critical for patients. Posted on 28 Sep 2021
A wide array of metagenomic study efforts are hampered by the same challenge: low concentrations of targets of interest combined with overwhelming amounts of background signal. Although PCR or naive DNA capture can be used when there are a small number of organisms of interest, design challenges become untenable for large numbers of targets.
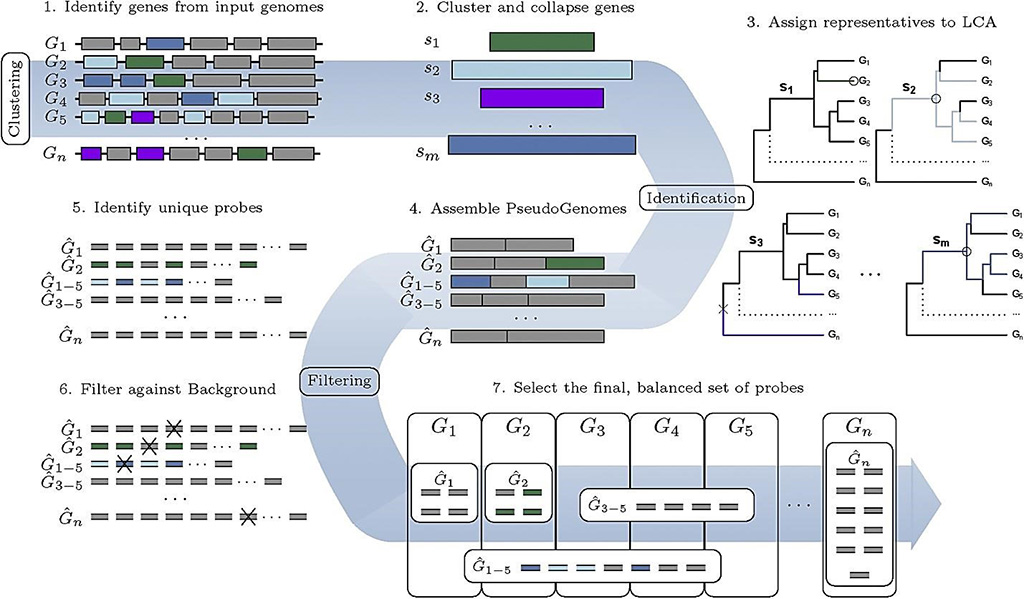
Image: The workflow of the HUBDesign pipeline: Probe design for simultaneous, targeted capture of diverse metagenomic targets (Photo courtesy of McMaster University)
A team of scientists at McMaster University (Hamilton, ON, Canada) and their colleagues have developed a sophisticated new tool that could help provide early warning of rare and unknown viruses in the environment and identify potentially deadly bacterial pathogens which cause sepsis. The new algorithm is an advanced tool that can help develop probes to capture trace quantities of pathogens, both known and unknown from a wide variety of situations, like the animal-to-human transmission of infections such as SARS-CoV-2 or monitor reservoirs in the environment for possible emerging pathogens.
The team successfully tested the probes on the entire family of coronaviruses, including SARS-CoV-2. The probes provide a shortcut by targeting, isolating and identifying the DNA sequences, specifically and simultaneously, that are shared among related organisms, most often due to evolutionary history or ancestry. To demonstrate the capabilities and effectiveness of the tool, The Hierarchical Unique Bait Design (HUBDesign), they designed and tested two probe sets: a coronavirus probe set capable of simultaneously detecting all sequenced coronaviruses, and a set of probes targeting bacterial pathogens associated with sepsis.
The majority (62.5%) of the probes have targets that are specific to one virus. Of the probes targeting multiple viruses, most (78.1%) target two or three. The remaining three sets of probes target loci specific to merbecoviruses and embecoviruses (both are Betacoronavirus subgenera) and loci common to the Deltacoronavirus genus. Both SARS-CoV-2 and HCoV-NL63 have probes at two levels in the hierarchy.
The HUBDesign probe set for sepsis pathogens contained 26,870 probes targeting bacterial pathogens, covering 2.09% of all nucleotides in the input dataset at an average depth of coverage of 3.64x. The investigators demonstrated the effectiveness of probes for capturing the incredibly wide array of pathogens associated with sepsis, a life-threatening and rapidly developing condition that occurs when the body overreacts to an infection which typically starts in the lungs, urinary tract, skin or gastrointestinal tract.
Hendrik Poinar, PhD, a Professor of Molecular Evolutionary Genetics and a lead author on the study, said, “We currently need faster, cheaper and more succinct ways to detect pathogens in human and environmental samples that democratize the hunt and this pipeline does exactly that.” The study was published on September 15, 2021 in the journal Cell Reports Methods.
Related Links:
McMaster University