CtDNA Analysis Assesses Early Relapse Detection in Colorectal Cancer
By LabMedica International staff writers
Posted on 17 Jun 2021
Colorectal cancer (CRC) is a type of cancer that begins in the large intestine (colon). The colon is the final part of the digestive tract. Colon cancer typically affects older adults, though it can happen at any age. It usually begins as small, noncancerous (benign) clumps of cells called polyps that form on the inside of the colon. Over time some of these polyps can become colon cancers.Posted on 17 Jun 2021
Challenges in the postoperative management of stage III CRC include selection of high-risk patients for adjuvant chemotherapy (ACT), lack of markers to assess ACT efficacy, assessment of recurrence risk after ACT, and, lack of markers to guide treatment decisions for high-risk patients e.g. additional therapy or intensified surveillance. Circulating tumor DNA (ctDNA) is a promising marker with potential to mitigate the challenges.
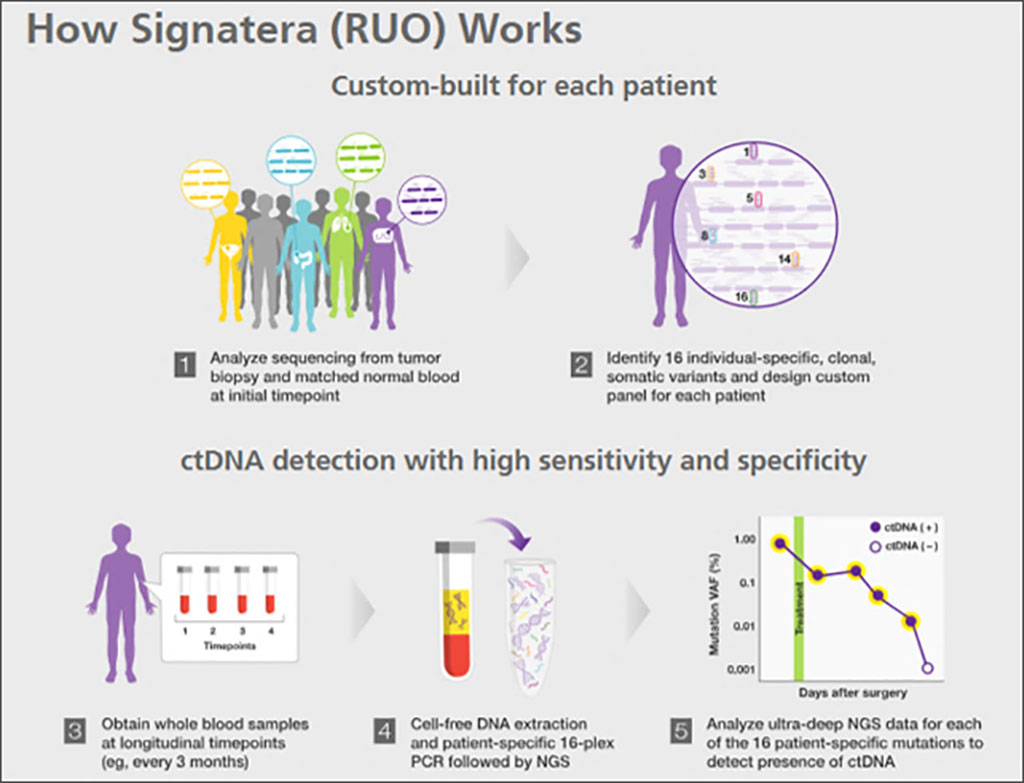
Image: Schematics of how the Signatera bespoke mPCR NGS assay works (Photo courtesy of Natera)
Medical Scientists from Aarhus University Hospital (Aarhus, Denmark) collected blood samples from 166 stage III CRC patients treated with curative intent at Danish and Spanish hospitals in 2014-2019. There were 1, 227 blood samples were collected prior to and immediately after surgery, and every third month for up to 36 months. Per patient 16 personal mutations were used to quantify plasma ctDNA, using the Signatera, bespoke mPCR NGS assay Natera, Austin, TX, USA).
The team reported that detection of ctDNA was a strong recurrence predictor, both postoperatively (Hazard ratio [HR] = 7.2), directly after ACT (HR = 18.2), and when measured serially after end of treatment (HR = 41). The recurrence rate of postoperative ctDNA positive patients treated with ACT was 80% (16/20). Patients who stayed ctDNA positive during ACT, all recurred. Serial post-treatment ctDNA measurements revealed exponential growth for all recurrence patients following either a SLOW (26% increase/month) or a FAST (126% increase/month) pattern.
From ctDNA detection to radiologic recurrence, ctDNA levels of FAST patients increased by a median 117-fold, and up to 554-fold. The 3-year overall survival was 43% for FAST patients and 100% for SLOW and non-recurrence patients (HR = 41.3). Coinciding CT scans and ctDNA measurements (n = 113 patients, 235 coinciding events, median 2 per patient) showed a high agreement (92%) and ctDNA either detected residual disease before the CT scan (n = 7 patients) or at the same time (n = 14 patients). The median lead-time was 7.5 months.
The authors concluded they had confirmed the prognostic power of serial postoperative ctDNA analysis. Moreover, it provided novel analyses demonstrating that ctDNA is more sensitive for recurrence detection than CT scans and can be used for tumor growth rate assessments. The difference between FAST and SLOW growing tumors suggest that growth rates could guide whom to start on systemic therapy rapidly and whom to send for diagnostic imaging. The study was presented at the 2021 ASCO Annual Meeting held June 4-8, 2021.
Related Links:
Aarhus University Hospital
Natera