Rapid Lateral Flow Assays Detect COVID-19 Variants and Differentiate COVID-19 from Other Respiratory Viral Diseases
By LabMedica International staff writers
Posted on 02 Mar 2021
A recent publication reported the development of two rapid diagnostic tests - one that detects COVID-19 variants and one that differentiates COVID-19 from other respiratory viral diseases.Posted on 02 Mar 2021
Investigators at the University of Minnesota Medical School (Minneapolis/St.Paul, USA) used the CRISPR/Cas9 gene editing tool to develop two rapid lateral flow diagnostic tests. CRISPRs (clustered regularly interspaced short palindromic repeats) are segments of prokaryotic DNA containing short repetitions of base sequences. Each repetition is followed by short segments of "spacer DNA" from previous exposures to a bacterial virus or plasmid. Since 2013, the CRISPR/Cas9 system has been used in research for gene editing (adding, disrupting, or changing the sequence of specific genes) and gene regulation. By delivering the Cas9 enzyme and appropriate guide RNAs (sgRNAs) into a cell, the organism's genome can be cut at any desired location. The conventional CRISPR/Cas9 system from Streptococcus pyogenes is composed of two parts: the Cas9 enzyme, which cleaves the DNA molecule and specific RNA guides that shepherd the Cas9 protein to the target gene on a DNA strand.
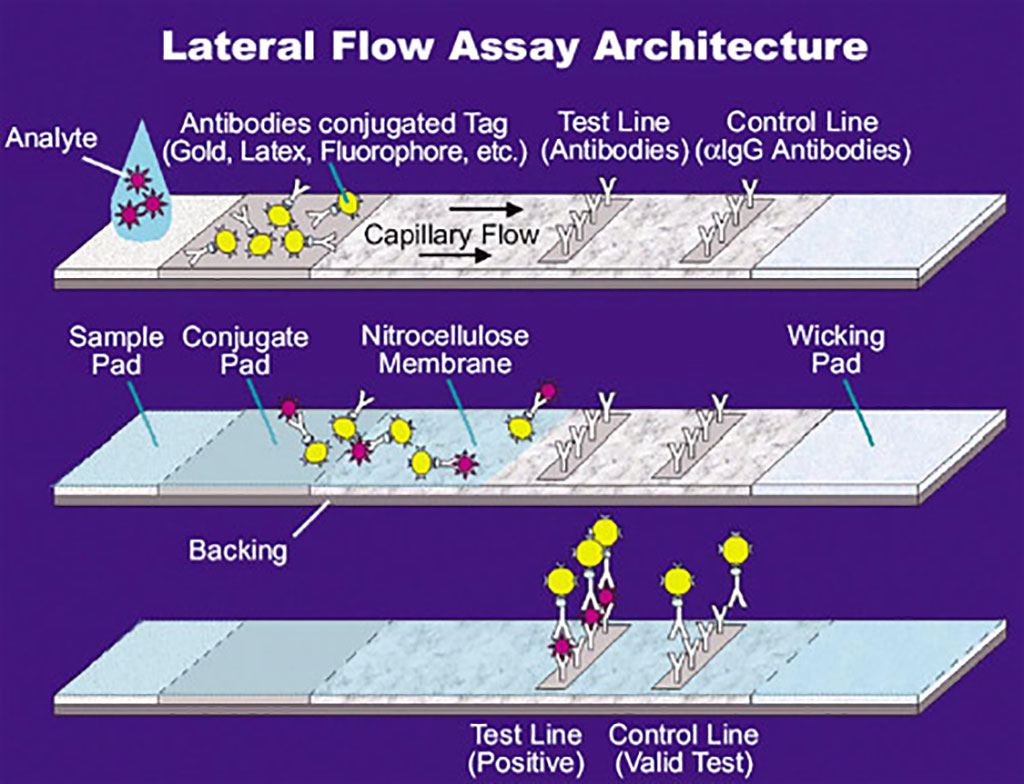
Image: Illustration of a lateral flow assay (LFA) (Photo courtesy of U.S. National Aeronautics and Space Administration via Wikimedia Commons)
The investigators integrated commercially available reagents into a CRISPR/Cas9-based lateral flow assay (LFA) that could detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) sequences with single-base specificity. This approach required minimal equipment and represented a simplified platform for field-based deployment. They also developed a rapid, multiplex fluorescence CRISPR/Cas9 nuclease cleavage assay capable of detecting and differentiating SARS-CoV-2, influenza A and B, and respiratory syncytial virus (RSV) in a single reaction.
The LFA test strips employed bound fluorescein isothiocyanate (FITC)/6-Carboxyfluorescein (FAM) and biotin to generate a positive result. Therefore, the investigators used a FITC/FAM-labeled PCR primer and a nuclease inactive (“dead”) biotinylated Cas9 and a single sgRNA specific for the ORF8a gene of SARS-Co-V-2 to label amplicons for detection by LFA. This approach was capable of single-nucleotide resolution and avoided false positives from primer dimer or non-specific amplification artifacts that could occur with the use of tandem FITC- and biotin-labeled primers for LFA.
"The approval of the SARS-CoV-2 vaccine is highly promising, but the time between first doses and population immunity may be months," said first author Dr. Mark J. Osborn, assistant professor of pediatrics at the University of Minnesota Medical School. "This testing platform can help bridge the gap between immunization and immunity."
The rapid LFA tests were described in the February 12, 2021, online edition of the journal Bioengineering.
Related Links:
University of Minnesota Medical School













